Difference between revisions of "FlyBase:Using FTP Archives"
m |
|||
| Line 8: | Line 8: | ||
| − | When viewed in a | + | When viewed in a browser, each FTP archive folder can be sorted by clicking on the column heading (Name, Last Modified, Size) multiple times to sort by alphabetical or reverse alphabetical order. |
There are multiple rendundant ways to reach the same file, so if two files in different folders have the exact same title, they are the same. | There are multiple rendundant ways to reach the same file, so if two files in different folders have the exact same title, they are the same. | ||
Latest revision as of 15:37, 22 February 2024
FlyBase is no longer supporting the archive servers, which replicated the website as it existed at the time of archiving. However, all of the data behind every release is available at our FTP archives, which you can reach on the Archived Data page, from the Downloads tab of the menu bar. This wiki page aims to help users become more familiar with these files, so they can figure out how to recapitulate searches done on the full website.
From the Archived Data page, scroll down to the Main Data Archives section to find links to the two main division of the FTP archive:
When viewed in a browser, each FTP archive folder can be sorted by clicking on the column heading (Name, Last Modified, Size) multiple times to sort by alphabetical or reverse alphabetical order.
There are multiple rendundant ways to reach the same file, so if two files in different folders have the exact same title, they are the same.
If you have trouble finding data within these files, please don't hesitate to contact FlyBase!
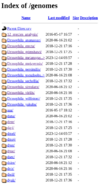
Genomes archive folder
The FTP genomes archive folder holds genomic sequence data from many Drosophilid species, organized in subsequent folders, first by species and then by release. If you are interested in a particular non-melanogaster Drosophilid, this is the easiest way to find all data specific to that species.
Data includes sequences in FASTA, GFF, and GTF file formats, as well as the Chado-XML database files for that species and release. The dna/ folders contain unprocessed sequences as .raw scaffold files.
The fullname (e.g. Drosophila_grimshawi) and four-letter FlyBase species abbreviation (e.g. Dgri) folders are different, but they contain identical duplicate files within them.
Precomputed files were never made for non-melanogaster species, so they are not available here.
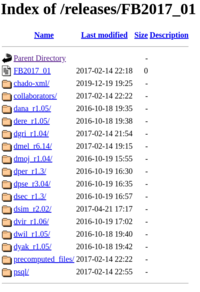
Release archive folder
The FTP releases archive folder holds data organized by release, for both D. melanogaster and non-melanogaster Drosophilids. These folders include:
- a chado-XML/ folder, containing the Chado-XML database files
- a collaborators/ folder, containing packages of data that we provide to other biological databases
- a folder for each species (e.g. dgri.../) containing genomic data (same files as above in the Genomes archive folder)
- a precomputed_files/ folder, containing precomputed files, which are plaintext tab-separated files that provide helpful intersections of FlyBase data types (e.g., fbal_to_fbgn_fb_*.tsv which lists all alleles associated with all genes).
- a psql/ folder, continaing compressed SQL files of the Chado database
Most queries, whether via with QuickSearch and a HitList, Batch Download, or Vocabularies are filtering information found in one of the Precomputed Files. Using the information in our wiki, you will hopefully be able to find a file that meets your needs.
You may need to do further filtering on these files, as they are large. Importing them as plain text into a spreadsheet program is a convenient way to search in them. Once the data is imported, you can use Find/searching, column sorting and filtering, or a pivot table to find the lines in that table containing your information of interest.