Difference between revisions of "FlyBase:CytoSearch Help"
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | =About CytoSearch= | |
| − | |||
A CytoSearch query will return lists of all the mapped genes, transgene insertions, and aberrations that are within, overlap or include the query region. The query returns features mapped both at the cytological and at the sequence level. Each hit includes the cytology, the observed (in green) or estimated (in red) sequence coordinates, and the symbol of the mapped feature as well as available stocks. | A CytoSearch query will return lists of all the mapped genes, transgene insertions, and aberrations that are within, overlap or include the query region. The query returns features mapped both at the cytological and at the sequence level. Each hit includes the cytology, the observed (in green) or estimated (in red) sequence coordinates, and the symbol of the mapped feature as well as available stocks. | ||
| Line 6: | Line 5: | ||
For CytoSearch searches, sequence-based data trumps cytology when both are available, cytology trumps meiotic data when both are available, and estimated cytology is used when only meiotic data are available. The FlyBase correspondence tables for cytological and sequence level maps are used to estimate cytology from sequence range and sequence range from cytology, for both the underlying data and the query input. | For CytoSearch searches, sequence-based data trumps cytology when both are available, cytology trumps meiotic data when both are available, and estimated cytology is used when only meiotic data are available. The FlyBase correspondence tables for cytological and sequence level maps are used to estimate cytology from sequence range and sequence range from cytology, for both the underlying data and the query input. | ||
| − | + | =Setting up a Search= | |
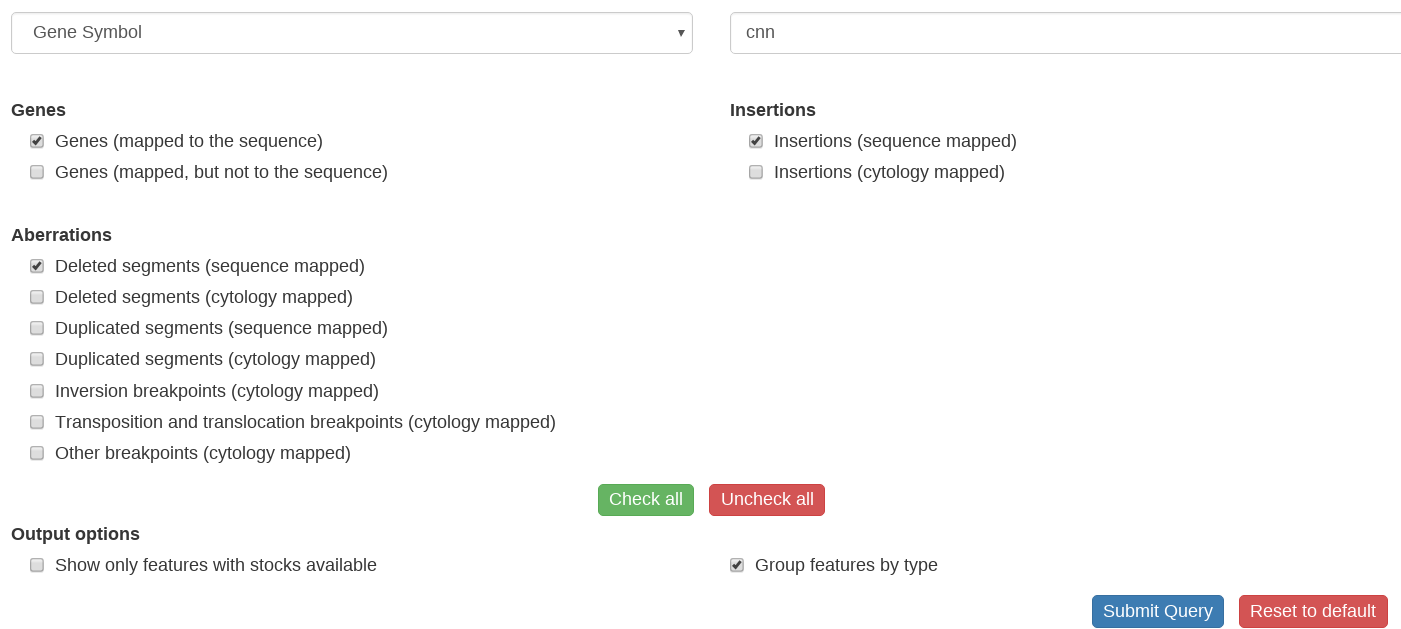
*Choose the type of search value you would like to use (cytological location, sequence coordinates, gene or annotation symbol or ID), from the dropdown menu. | *Choose the type of search value you would like to use (cytological location, sequence coordinates, gene or annotation symbol or ID), from the dropdown menu. | ||
| Line 14: | Line 13: | ||
*Click on the “Submit Query” button. | *Click on the “Submit Query” button. | ||
| − | + | =Example CytoSearch search (picture only, not live)= | |
| − | |||
[[File:CytoSearch_cnn.png]] | [[File:CytoSearch_cnn.png]] | ||
| − | + | =Features of Results Page= | |
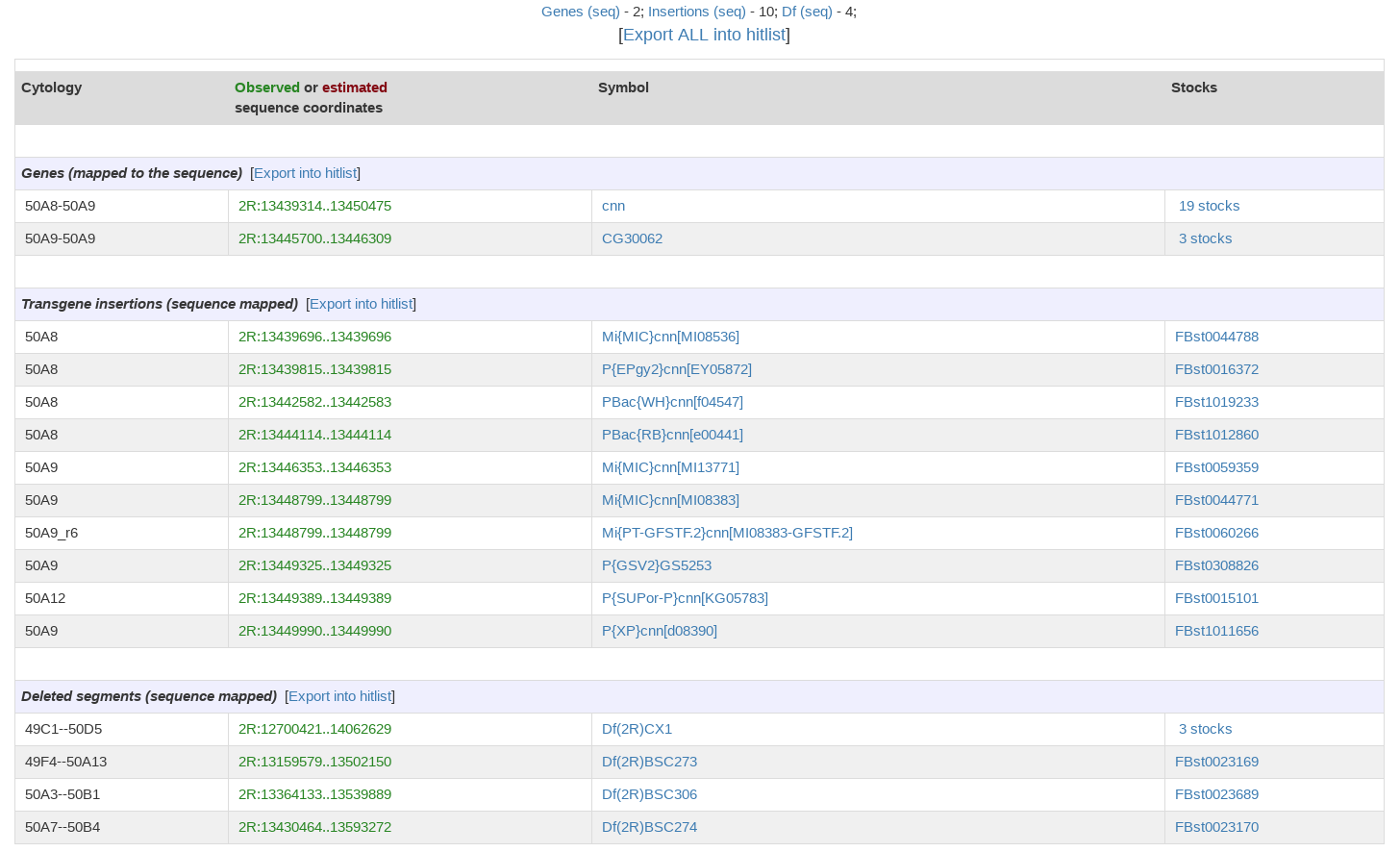
*The number of features of each type are displayed across the top of the results page. | *The number of features of each type are displayed across the top of the results page. | ||
| − | *The sequence ranges on the results page link to ''' | + | *The sequence ranges on the results page link to '''JBrowse''', which provides a graphical display of annotation, insertion, and aberration data for that region. |
*Gene, insertion, aberration, and stock symbols on the results page link to their respective FlyBase reports. If multiple stocks are available, the number of stocks is shown, which links to a hit list of relevant stocks. | *Gene, insertion, aberration, and stock symbols on the results page link to their respective FlyBase reports. If multiple stocks are available, the number of stocks is shown, which links to a hit list of relevant stocks. | ||
| Line 28: | Line 26: | ||
*Data on the results page can be '''exported''' to Batch Download or QueryBuilder by clicking on the blue “Export ALL into hitlist” box above the results table. If you wish to export the results for one data type, choose the option to sort the CytoSearch results by type when you run the search, then choose the “Export into hitlist” option for your data type of interest. You will then have access to the many options available from the hit list pages. These are described on the [hitlist help page] (link). | *Data on the results page can be '''exported''' to Batch Download or QueryBuilder by clicking on the blue “Export ALL into hitlist” box above the results table. If you wish to export the results for one data type, choose the option to sort the CytoSearch results by type when you run the search, then choose the “Export into hitlist” option for your data type of interest. You will then have access to the many options available from the hit list pages. These are described on the [hitlist help page] (link). | ||
| − | + | =Results Page for the Sample Search (picture only, not live)= | |
| − | |||
| − | |||
[[File:CytoSearch_result_cnn.png]] | [[File:CytoSearch_result_cnn.png]] | ||
Latest revision as of 17:48, 20 December 2022
About CytoSearch
A CytoSearch query will return lists of all the mapped genes, transgene insertions, and aberrations that are within, overlap or include the query region. The query returns features mapped both at the cytological and at the sequence level. Each hit includes the cytology, the observed (in green) or estimated (in red) sequence coordinates, and the symbol of the mapped feature as well as available stocks.
For CytoSearch searches, sequence-based data trumps cytology when both are available, cytology trumps meiotic data when both are available, and estimated cytology is used when only meiotic data are available. The FlyBase correspondence tables for cytological and sequence level maps are used to estimate cytology from sequence range and sequence range from cytology, for both the underlying data and the query input.
Setting up a Search
- Choose the type of search value you would like to use (cytological location, sequence coordinates, gene or annotation symbol or ID), from the dropdown menu.
- Enter the search value in the box to the right.
- Select the classes of mapped entities you want returned in the search from the list below grouped into Genes, Insertions, and Aberrations options. The default search will list all features in the region sorted by cytological location.
- Check options to return only features with available stocks or to group the results by type if desired.
- Click on the “Submit Query” button.
Example CytoSearch search (picture only, not live)
Features of Results Page
- The number of features of each type are displayed across the top of the results page.
- The sequence ranges on the results page link to JBrowse, which provides a graphical display of annotation, insertion, and aberration data for that region.
- Gene, insertion, aberration, and stock symbols on the results page link to their respective FlyBase reports. If multiple stocks are available, the number of stocks is shown, which links to a hit list of relevant stocks.
- Data on the results page can be exported to Batch Download or QueryBuilder by clicking on the blue “Export ALL into hitlist” box above the results table. If you wish to export the results for one data type, choose the option to sort the CytoSearch results by type when you run the search, then choose the “Export into hitlist” option for your data type of interest. You will then have access to the many options available from the hit list pages. These are described on the [hitlist help page] (link).