FlyBase:Author Reagent Table (ART)
Rationale and Purposes
Wider use of identifiers and recognized symbols increases the transparency and reproducibility of biological research, while aiding curation into research databases.
The table is designed to be used regularly during the course of a research project, recording reagents as they are received and/or used. Lab-wide use of a common reagent form would facilitate tracking of reagents within the lab.
A spreadsheet format is convenient and flexible for the researcher and provides commonly used download options for readers of publications. Use of the same format throughout (reagent data collection, submission for publication, post-publication downloads) reduces error-prone copying and pasting steps.
Providing information in a structured format that allows bulk downloads greatly facilitates curation into research databases.
In addition to reagents, unambiguous identification of specific genes studied is particularly helpful for genetic and genome databases, as well as for the larger research community.
Download the ART template
Download template in XLSX format 
|
Guidelines for creation of the Author Reagent Table
These guidelines are also available in .docx format.
Some of the constraints below are imposed to allow automated parsing of the table.
Columns (general):
- Column order and headings must be preserved.
- Columns A-E should not be removed, even if not used.
- Additional columns can be appended (F, G, etc.) for internal use.
Rows (general):
- Row titles (preceding the parentheses) must be preserved.
- Row one (column headings) should not be removed.
- Row two (headings with instructions) may be removed.
- Order of the rows can be changed.
- Clustering of multiple reagents of one type recommended, but is not mandatory.
- Colors are entirely optional: can be removed or changed.
- Data types indicated on template that are not used can be removed.
- Data types not represented in the template: use “other”.
Column A: Reagent type (mandatory)
- Row title (preceding parentheses) cannot be changed.
- See descriptions of categories in “Reagent categories” section below.
- If appropriate, include species within parentheses. Use full genus and full species for first mention; thereafter, initial for genus may be used.
- If appropriate, indicate sex after species (within the parentheses); separate by comma, space.
- See additional information in the section “Rows (general)” above.
Column B: Designation used in publication (mandatory)
- This is a free-text field; there are no formatting constraints, except for separation of multiple entries by semi-colon, space.
- Should indicate exactly how the reagent is referred to in the publication. If several designations are used for one reagent, can include more than one entry, separated by semi-colon, space. If one designation has been used to refer to several different reagents, list each reagent separately and include a unique identifier or description parenthetically.
- If there is a semi-colon within the designation used, add double quotes around the entire phrase.
- Additional defining information can be included parenthetically (if brief).
Column C: Source or reference (mandatory)
- If reagent obtained from a public source, such as a stock center or company, list resource name.
- If not obtained from public source, but a published description is available, list the DOI or PMID for the publication.
- If reagent newly created in the publication, enter “this paper”.
- If none of the above, enter “other” and explain in Additional Information (Column E).
- If it is appropriate to indicate both a publication and a lab or researcher that provided the reagent, list the publication in this field and the lab or researcher in Additional Information.
Column D: Identifiers (not mandatory; use if possible)
- Use the format “ID_source:identifier” with a colon and no spaces. Separate multiple entries with a semi-colon, space.
- PLEASE DO NOT GUESS. If you are not certain, leave column D blank and enter in the ‘Additional Information’ (column E) any pertinent information about the origin of the reagent (e.g. laboratory from which reagent was obtained).
- If possible, use the recommended abbreviation for the ID_source. These are included in the downloadable ART Guidelines, .docx format.
- Multiple identifiers, even if redundant, are useful for validation. In some case a more general identifier (for a stock, for instance) can be combined with a more precise identifier (for the allele of interest in that stock).
- Inclusion of database identifiers is strongly encouraged to facilitate accurate incorporation of published data into appropriate biological databases.
Column E: Additional information
- Mandatory if column C entry is “this paper” or “other”. Description of new reagent or laboratory from which reagent obtained. (Free text)
- For antibodies, indicate dilution in parentheses (to avoid auto-formatting problems).
- Other pertinent information, including pointers to supplementary information or Methods section, if appropriate. (Free text)
Additional columns can be added for internal purposes (and removed prior to submission for publication): tracking acquisition by the lab, lab-specific stock numbers, storage location in the lab, comments about problems or QC, etc.
Reagent types
- Gene (indicate species) - not strictly a reagent, but unambiguous identification critical for genetic research and genetic databases.
- Strain, strain background (indicate species) - applies to whole organism; includes bacterial and virus strains or isolates. Indicate sex, if applicable.
- Genetic reagent (indicate species) - applies to mutations and variants in whole organism, including transgenically introduced constructs. For transgenic lines, indicate host species. Indicate sex, if applicable.
- Cell line (indicate species) - if a primary cell line, describe in Additional Information. Indicate sex, if applicable.
- Transfected construct (indicate species) - in cell line. Generally, indicate species of cell line; use species of construct component if that is more relevant and explain in Additional Information.
- Biological sample (indicate species) - any other biological entity, ranging from isolated tissue to defined population; describe in Additional Information. Indicate sex, if applicable.
- Antibody - include host organism common name and clonality (e.g., “mouse monoclonal”); include dilution used in Additional Information (within parentheses to avoid auto-formatting problems).
- Recombinant DNA reagent - traditional cultured clones, plasmids, cDNAs, etc., including recombinant DNA libraries.
- Sequence-based reagent - oligonucleotides, primers, morpholinos, etc.; indicate sequence.
- Peptide, recombinant protein - generally, commercially available reagents.
- Chemical compound, drug - generally, commercially available reagents.
- Commercial assay, kit - detection assays; labeling and sample preparation kits.
- Large-scale dataset - newly created or previously existing; include data repository accession number(s) in Identifiers column.
- Software, algorithm - newly created or previously existing.
- Other - miscellaneous other categories, including histological stains.
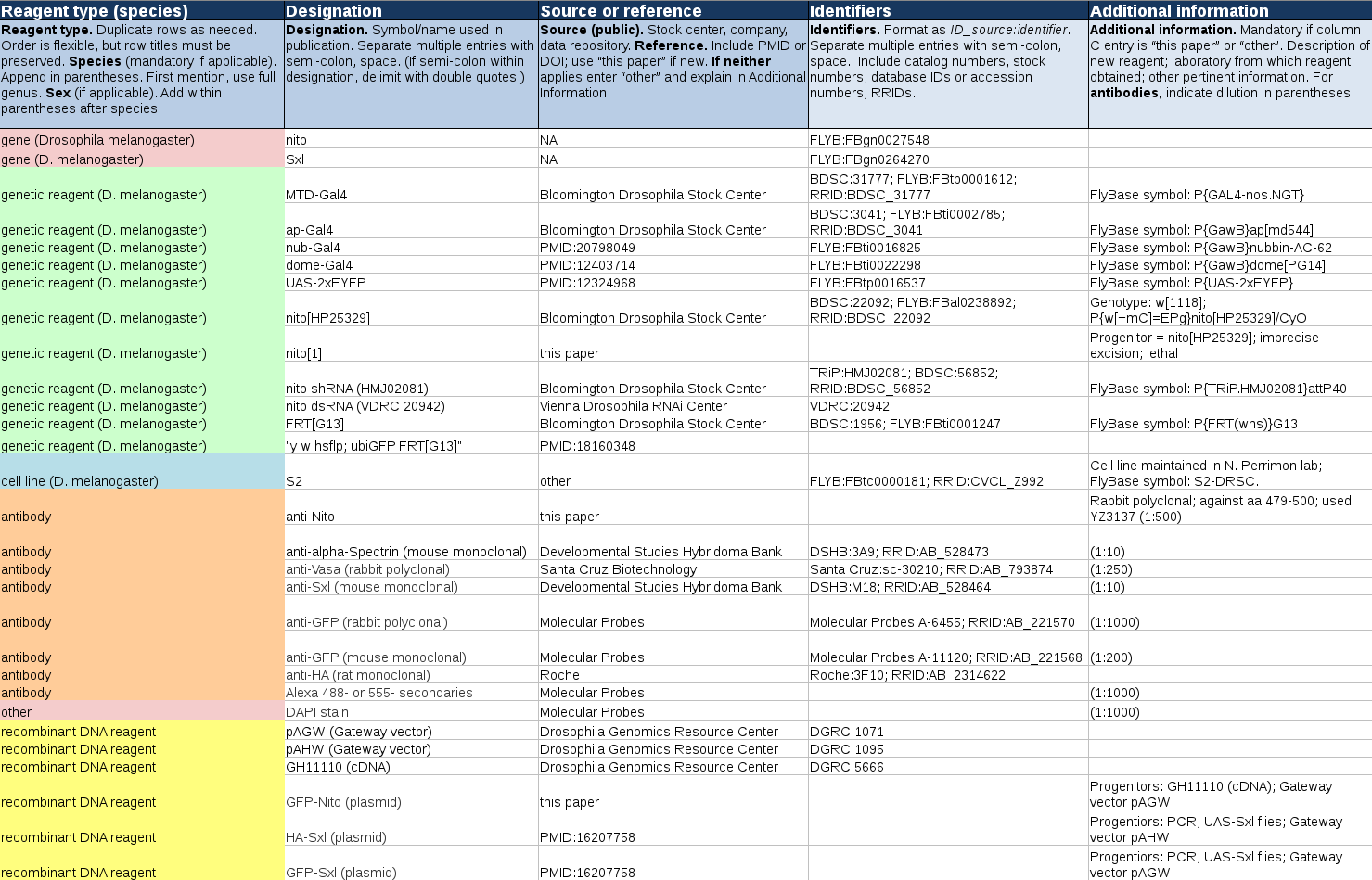
Example of Completed Author Reagent Table
Download an xlsx version of the example below.
List of ID_source abbreviations
Additional entries and improvements solicited. Have used a minimum of 3 letters to minimize potential conflicts.
Companies: Full name should be entered in column C; use first two words as identifier prefix (or one, if single word). Do not simply indicate "Catalog #" -- this is too error-prone. Databases: