Difference between revisions of "FlyBase:Fast-Track Your Paper"
Aoife Larkin (talk | contribs) |
Aoife Larkin (talk | contribs) |
||
| (22 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
==Overview== | ==Overview== | ||
| − | The [http://{{flybaseorg}}/submission/publication/ Fast-Track Your Paper] (FTYP) tool allows first-pass curation by users, indicating to curators the types of data in the paper and also resulting in relevant genes being associated with the reference. Corresponding authors of new publications are emailed and asked to use FTYP when their paper has been fully published (''i.e.'' has final volume and page numbers) and has been added to our database. The Fast-Track Your Paper tool may also be accessed via the ‘Tools’ menu in the navigation bar present on all FlyBase pages. | + | The [http://{{flybaseorg}}/submission/publication/ Fast-Track Your Paper] (FTYP) tool allows first-pass curation by users, indicating to curators the types of data in the paper and also resulting in relevant genes being associated with the reference. Corresponding authors of new publications are emailed and asked to use FTYP when their paper has been fully published (''i.e.'' has final volume and page numbers) and has been added to our database. The Fast-Track Your Paper tool may also be accessed via the ‘Tools’ menu in the navigation bar present on all FlyBase pages. Please note, there will be a delay before data related to your FTYP submission (e.g. gene-reference associations) will be visible on the website (see the [https://{{flybaseorg}}/static/release_schedule FlyBase release schedule]); FlyBase curators will immediately be able to access your FTYP submission in order to prioritize curation. |
==Quick instructions== | ==Quick instructions== | ||
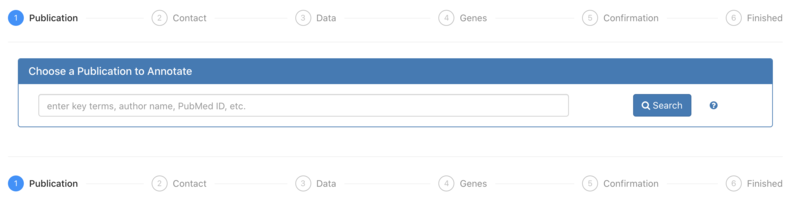
1. Authors coming to FTYP from an email to alert them about their publication will not need to complete this Publication step, as their paper will already have been selected. Otherwise, users may search for a particular publication using keywords. | 1. Authors coming to FTYP from an email to alert them about their publication will not need to complete this Publication step, as their paper will already have been selected. Otherwise, users may search for a particular publication using keywords. | ||
| − | [[File:FTYP_1a._Publication_step.png|thumb| | + | [[File:FTYP_1a._Publication_step.png|thumb|center|800px|Publication search]] |
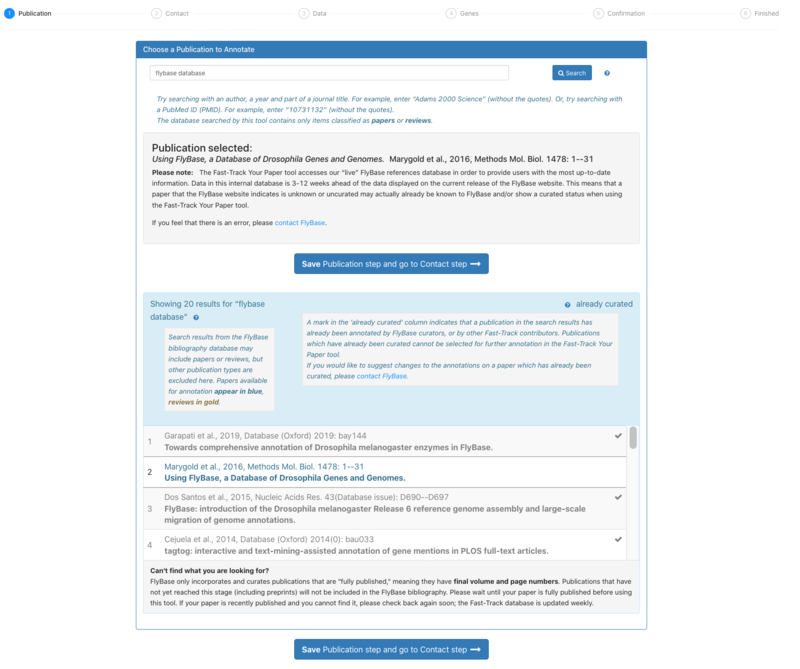
2. Publications available for the FTYP process are in blue (papers) or gold (reviews). Those papers that have already been curated appear in grey and are not selectable. | 2. Publications available for the FTYP process are in blue (papers) or gold (reviews). Those papers that have already been curated appear in grey and are not selectable. | ||
| − | [[File:FTYP_1b._Publication_step.png|thumb| | + | [[File:FTYP_1b._Publication_step.png|thumb|center|800px|Publication step]] |
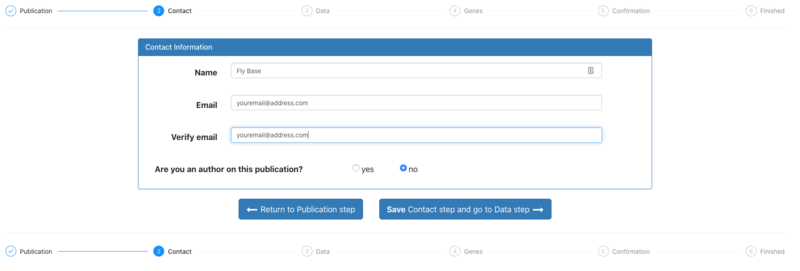
3. Authors coming to FTYP from an email to alert them about their publication will have their email address filled in already in the Contact step. Otherwise, users should fill in their name and email address and indicate if they are an author on the publication. | 3. Authors coming to FTYP from an email to alert them about their publication will have their email address filled in already in the Contact step. Otherwise, users should fill in their name and email address and indicate if they are an author on the publication. | ||
| − | [[File:FTYP_2._Contact_step.png|thumb| | + | [[File:FTYP_2._Contact_step.png|thumb|center|800px|Contact step]] |
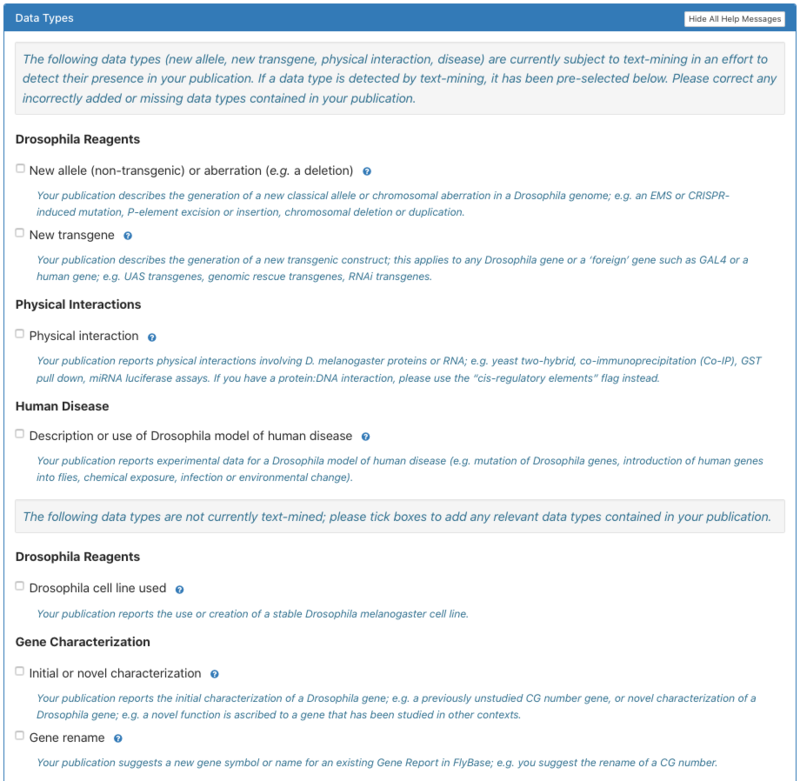
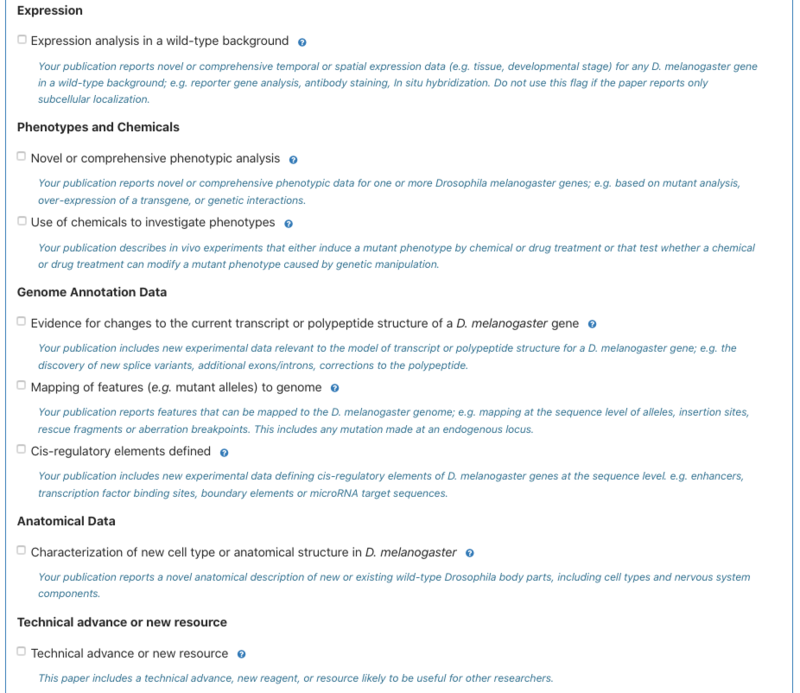
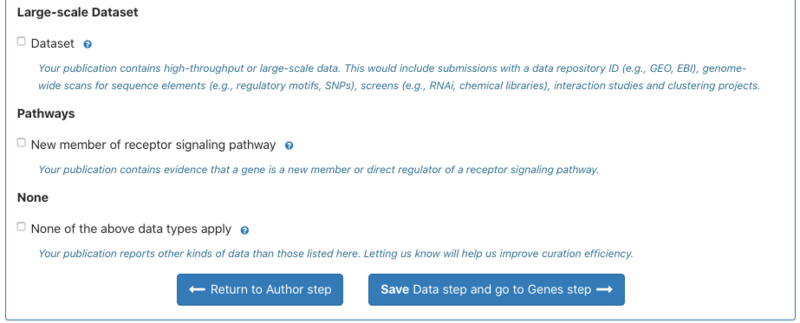
4. Users can indicate the types of data the paper contains in the Data step; if curating a review the user will not encounter this step. Some data-types (new allele, new transgene, physical interaction, human disease) may be pre-populated based on text-mining, but can be corrected by the user. Users should tick any data types that apply to their paper. If none apply, please tick the ‘none of the above data types apply’ box and optionally tell us the kind of data that is in your paper. | 4. Users can indicate the types of data the paper contains in the Data step; if curating a review the user will not encounter this step. Some data-types (new allele, new transgene, physical interaction, human disease) may be pre-populated based on text-mining, but can be corrected by the user. Users should tick any data types that apply to their paper. If none apply, please tick the ‘none of the above data types apply’ box and optionally tell us the kind of data that is in your paper. | ||
| − | [[File: | + | [[File:FTYP_data_top.png|thumb|center|800px]] |
| + | [[File:FTYP_data_middle.png|thumb|center|800px]] | ||
| + | [[File:FTYP_data_bottom.png|thumb|center|800px]] | ||
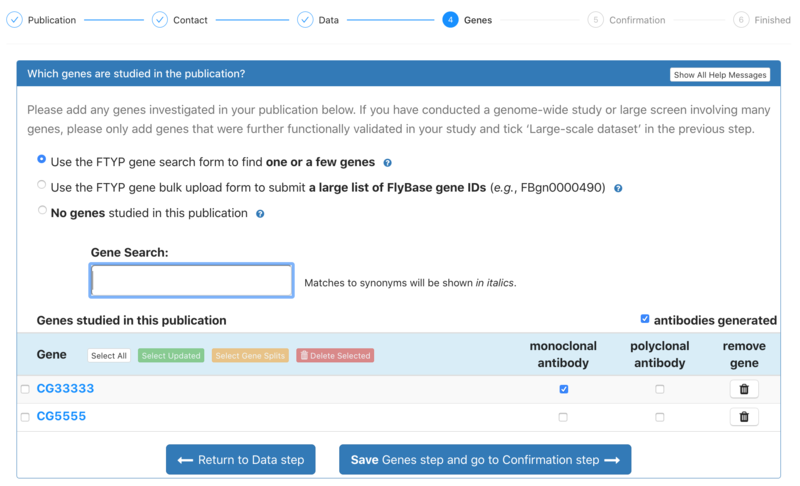
| − | 5. During the Gene step, users should associate those genes that were studied in the publication. These can be added manually one by one, using the Gene Search box. Alternatively, a larger list of genes (no more than 100) can be added in the form of a list of FlyBase gene IDs (e.g. FBgn0000490). However, papers including large screens or genome-wide studies that involve many genes should only add genes that were functionally validated in the study, and tick the ‘Large-scale dataset’ box in the previous Data step. The user can also indicate if any antibodies were generated for particular genes. There is the option to specify that no genes were studied in the publication. | + | 5. During the Gene step, users should associate those genes that were studied in the publication. These can be added manually one by one, using the Gene Search box. Alternatively, a larger list of genes (no more than 100) can be added in the form of a list of FlyBase gene IDs (''e.g.'' FBgn0000490). However, papers including large screens or genome-wide studies that involve many genes should only add genes that were functionally validated in the study, and tick the ‘Large-scale dataset’ box in the previous Data step. The user can also indicate if any antibodies were generated for particular genes. There is the option to specify that no genes were studied in the publication. |
| − | [[File:FTYP_4._Gene_step.png|thumb| | + | [[File:FTYP_4._Gene_step.png|thumb|center|800px|Gene step]] |
| + | |||
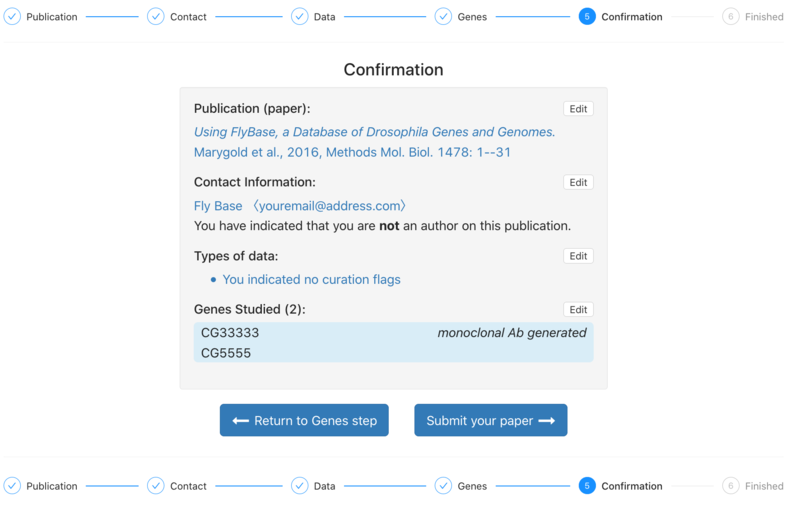
| + | 6. The Confirmation step shows a summary of the information associated with the paper. The user can still go back at this stage and edit information from previous steps. Once the user is happy with the attached information, the paper can be submitted. | ||
| + | [[File:FTYP_5._Confirmation_step.png|thumb|center|800px|Confirmation step]] | ||
| + | |||
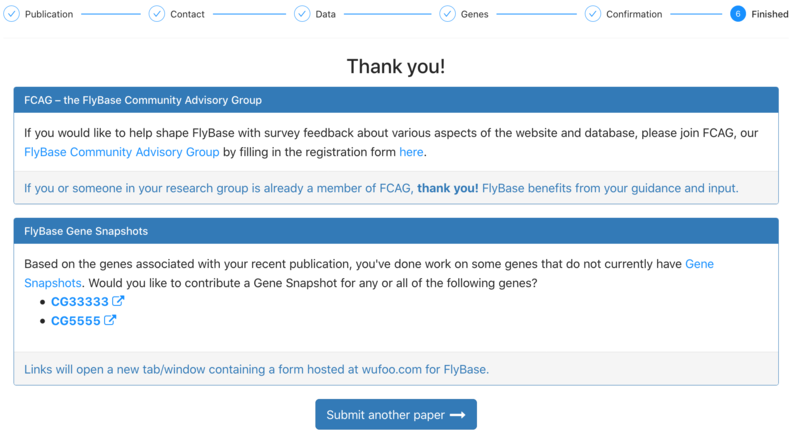
| + | 7. Thank you! After submission, the user is encouraged to help FlyBase in other ways: for example, by joining the FlyBase Community Advisory Group or by adding Gene Snapshots to genes studied in the paper that do not currently have a snapshot. They can also submit another paper using FTYP. | ||
| + | [[File:Post_submission.png|thumb|center|800px|Post submission]] | ||
Latest revision as of 14:21, 17 May 2022
Fast-Track Your Paper
Overview
The Fast-Track Your Paper (FTYP) tool allows first-pass curation by users, indicating to curators the types of data in the paper and also resulting in relevant genes being associated with the reference. Corresponding authors of new publications are emailed and asked to use FTYP when their paper has been fully published (i.e. has final volume and page numbers) and has been added to our database. The Fast-Track Your Paper tool may also be accessed via the ‘Tools’ menu in the navigation bar present on all FlyBase pages. Please note, there will be a delay before data related to your FTYP submission (e.g. gene-reference associations) will be visible on the website (see the FlyBase release schedule); FlyBase curators will immediately be able to access your FTYP submission in order to prioritize curation.
Quick instructions
1. Authors coming to FTYP from an email to alert them about their publication will not need to complete this Publication step, as their paper will already have been selected. Otherwise, users may search for a particular publication using keywords.
2. Publications available for the FTYP process are in blue (papers) or gold (reviews). Those papers that have already been curated appear in grey and are not selectable.
3. Authors coming to FTYP from an email to alert them about their publication will have their email address filled in already in the Contact step. Otherwise, users should fill in their name and email address and indicate if they are an author on the publication.
4. Users can indicate the types of data the paper contains in the Data step; if curating a review the user will not encounter this step. Some data-types (new allele, new transgene, physical interaction, human disease) may be pre-populated based on text-mining, but can be corrected by the user. Users should tick any data types that apply to their paper. If none apply, please tick the ‘none of the above data types apply’ box and optionally tell us the kind of data that is in your paper.
5. During the Gene step, users should associate those genes that were studied in the publication. These can be added manually one by one, using the Gene Search box. Alternatively, a larger list of genes (no more than 100) can be added in the form of a list of FlyBase gene IDs (e.g. FBgn0000490). However, papers including large screens or genome-wide studies that involve many genes should only add genes that were functionally validated in the study, and tick the ‘Large-scale dataset’ box in the previous Data step. The user can also indicate if any antibodies were generated for particular genes. There is the option to specify that no genes were studied in the publication.
6. The Confirmation step shows a summary of the information associated with the paper. The user can still go back at this stage and edit information from previous steps. Once the user is happy with the attached information, the paper can be submitted.
7. Thank you! After submission, the user is encouraged to help FlyBase in other ways: for example, by joining the FlyBase Community Advisory Group or by adding Gene Snapshots to genes studied in the paper that do not currently have a snapshot. They can also submit another paper using FTYP.