Difference between revisions of "Resource Portal"
ChrisTabone (talk | contribs) (→CRISPR) |
ChrisTabone (talk | contribs) |
||
| (207 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | < | + | FlyBase maintains extensive lists of useful external Network and Material Resources for fly researchers that can be accessed from the following links:<br /> |
| + | '''[http://flybase.org/wiki/FlyBase:Drosophila_Network_Resources Network Resources]'''<br/> | ||
| + | '''[http://flybase.org/wiki/FlyBase:Drosophila_Material_Resources Reagent Resources]'''<br/> | ||
| + | Links relevant to some of our most popular resources have been compiled and are listed below. | ||
= Popular Resources = | = Popular Resources = | ||
| − | {|class="wikitable | + | {|class="wikitable" |
|- | |- | ||
| − | |style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#CRISPR|CRISPR]]</big> ||style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#RNAi|RNAi]]</big> ||style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#Stock_Centers|Stock Centers]]</big> | + | |style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#CRISPR|CRISPR]]</big> ||style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#RNAi|RNAi]]</big> ||style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#Stock_Centers|Stock Centers]]</big> ||style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#Antibodies|Antibodies]]</big>||style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#Orthologs|Orthologs]]</big>||style="text-align: center; padding: 20px;"| <big>[[Resource_Portal#All_Resources|All Resources]]</big> |
|} | |} | ||
| − | == | + | == Full Table of Contents == |
| − | + | __TOC__ | |
| − | + | == CRISPR == | |
=== gRNA Design Resources === | === gRNA Design Resources === | ||
| Line 18: | Line 21: | ||
{|cellpadding=5 | {|cellpadding=5 | ||
|- | |- | ||
| − | !style="background: #efefef;"| Resource !!style="background: #efefef;"| | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source |
|- | |- | ||
| − | |style="background: #efefef;"| [http:// | + | |style="white-space: nowrap; background: #efefef;"| [http://tools.flycrispr.molbio.wisc.edu/targetFinder/ CRISPR Optimal Target Finder] ||style="background: #efefef;"| Identifies gRNA targets within a provided sequence and additionally searches genome-wide ('''release 6, current FlyBase release''') for potential off-target sites. Includes numerous <em>Drosophila</em> species.||style="white-space: nowrap; background: #efefef;"|O'Connor-Giles, Wildonger, and Harrison Labs<br /> University of Wisconsin-Madison<br /> WI, USA |
|- | |- | ||
| − | + | | [http://www.crisprscan.org/ CRISPRscan] || Searchable and browsable collection of genome-wide ('''release 6, current FlyBase release''') sgRNA target sites. Displays sgRNA sites as tracks on UCSC's genome browser and allows searching via gene or prediction via sequence. Support for numerous species (fly, zebrafish, mouse, human, others).|| Giraldez Lab<br /> Yale University<br /> New Haven, CT, USA | |
|- | |- | ||
| − | |style="background: #efefef;"| [http://www. | + | |style="background: #efefef;"|[http://www.flyrnai.org/crispr2/ DRSC Find CRISPR Tool] ||style="background: #efefef;"| A tool for discovering gRNA targets located throughout the fly genome (release 5) using gene IDs, symbols, or chromosome locations. Allows for filtering of off-target sites and control of mis-match stringency. ||style="background: #efefef;"| Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA |
|- | |- | ||
| − | | [ | + | | [http://www.e-crisp.org/E-CRISP/designcrispr.html E-CRISPR] || gRNA genome-wide (release 5) target finder, searchable via gene symbol or sequence. Support for numerous species (fly, zebrafish, mouse, human, others). Offers "relaxed, medium and strict" search options. ||Boutros lab, DKFZ<br/> Heidelberg, Germany |
|- | |- | ||
| − | |style="background: #efefef;"| [ | + | |style="background: #efefef;"| [https://chopchop.rc.fas.harvard.edu/ CHOPCHOP] ||style="background: #efefef;"| Search for gRNA targets (release 5). Flexible inputs (gene name, genomic coordinates or DNA sequence) and detection of off-target sites. ||style="background: #efefef;"| Schier & Church Labs<br /> Harvard University<br /> Cambridge, MA, USA |
|- | |- | ||
| − | | [http:// | + | | [http://crispr.mit.edu/ CRISPR Design] ||Identification of gRNA sites genome wide (release 5). Offers options for multiple species. || Zhang Lab<br /> MIT<br /> Boston, MA, USA |
|- | |- | ||
| − | |style="background: #efefef;"| [http://tefor.net/crispor/crispor.cgi CRISPOR] ||style="background: #efefef;"| | + | |style="background: #efefef;"| [http://tefor.net/crispor/crispor.cgi CRISPOR] ||style="background: #efefef;"| A program that helps design, evaluate, and clone guide sequences for the CRISPR/Cas9 system. Allows for searches across multiple species of <em>Drosophila</em>, including melanogaster (release 5).||style="background: #efefef;"| Tefor<br />France |
|- | |- | ||
| − | |||
|} | |} | ||
| + | <br/> | ||
=== CRISPR Stocks === | === CRISPR Stocks === | ||
| Line 41: | Line 44: | ||
{|cellpadding=5 | {|cellpadding=5 | ||
|- | |- | ||
| − | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Author !!style="background: #efefef;"| Additional | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source |
| + | |- | ||
| + | |style="white-space: nowrap; background: #efefef;"| [http://flystocks.bio.indiana.edu/Browse/misc-browse/CRISPR.htm CRISPR system stocks for site-specific mutagenesis]||style="background: #efefef;"| The Bloomington Stock Center's CRISPR/Cas9 page. Includes various stocks for expression of both Cas9 and tracrRNA. ||style="background: #efefef;"| Bloomington Drosophila Stock Center (BDSC)<br/> Indiana University<br/> Bloomington, IN, USA | ||
| + | |- | ||
| + | | [http://www.crisprflydesign.org/flies/ CRISPR Fly Design] || Information on stocks generated by the CRISPR Fly Design group at the MRC Laboratory of Molecular Biology. ||style="white-space: nowrap;"| MRC Laboratory of Molecular Biology<br /> | ||
| + | Cambridge, UK | ||
| + | |- | ||
| + | |} | ||
| + | <br/> | ||
| + | |||
| + | === CRISPR Vectors === | ||
| + | |||
| + | {|cellpadding=5 | ||
| + | |- | ||
| + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source | ||
| + | |- | ||
| + | |style="white-space: nowrap; background: #efefef;"| [https://www.addgene.org/crispr/drosophila/ Addgene CRISPR/Cas9 Plasmids for use in insects]||style="white-space: nowrap; background: #efefef;"| A list of available Cas9 & gRNA expression plasmids for use in insects, including <em>Drosophila</em>. ||style="background: #efefef;"| Addgene<br />Cambridge, MA, USA | ||
| + | |- | ||
| + | | [http://www.crisprflydesign.org/plasmids/ CRISPR Fly Design Plasmids] || Descriptions and information regarding the plasmids created by the CRISPR Fly Design group (also [http://www.addgene.org/search/advanced/?q=Bullock#p=true deposited at Addgene]). || MRC Laboratory of Molecular Biology<br /> | ||
| + | Cambridge, UK | ||
| + | |} | ||
| + | <br/> | ||
| + | === Additional Useful Links === | ||
| + | {|cellpadding=5 | ||
| + | |- | ||
| + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source | ||
| + | |- | ||
| + | |style="white-space: nowrap; background: #efefef;"| [http://www.crisprflydesign.org/ CRISPR Fly Design]||style="white-space: nowrap; background: #efefef;"| An overview and resource collection for CRISPR gene editing in <em>Drosophila</em>. ||style="background: #efefef;"| MRC Laboratory of Molecular Biology<br /> | ||
| + | Cambridge, UK | ||
| + | |- | ||
| + | | [http://flycrispr.molbio.wisc.edu/ flyCRISPR] || An overview and resource collection for CRISPR gene editing in <em>Drosophila</em>. || O'Connor-Giles, Wildonger, and Harrison Labs<br /> University of Wisconsin-Madison<br /> WI, USA | ||
| + | |- | ||
| + | | style="background: #efefef;"| [http://igtrcn.org/ Insect Genomic Technologies Research Coordination Network] || style="background: #efefef;"| Discussion and dissemination of advanced technologies for genome modification of insects through symposia, technical workshops, and training fellowships. || style="background: #efefef; white-space: nowrap;"|Institute for Bioscience and Biotechnology Research<br />University of Maryland<br />Rockville, MD, USA | ||
| + | |- | ||
| + | | [http://flyrnai.blogspot.com/search/label/CRISPRs FlyRNAi Blog, CRISPR tags] || A research blog tracking functional genomics techniques in <em>Drosophila</em>, filtered for keyword: CRISPRs. || Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.flyrnai.org/DRSC-Protocol_single_cell_cloning.html DRSC Single Cell Cloning Protocol] ||style="background: #efefef;"| Single-cell cloning method useful for <em>Drosophila</em> cell CRISPR gene editing ||style="background: #efefef;"| Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA | ||
| + | |} | ||
| + | <br/> | ||
| + | |||
| + | === Selected recent methods reviews === | ||
| + | |||
| + | * Venken KJ, Sarrion-Perdigones A, Vandeventer PJ, Abel NS, Christiansen AE, Hoffman KL. [http://www.ncbi.nlm.nih.gov/pubmed/26447401 '''Genome engineering: Drosophila melanogaster and beyond.'''] Wiley Interdiscip Rev Dev Biol. 2015 Oct 8. doi: 10.1002/wdev.214. [Epub ahead of print<nowiki>]</nowiki> Review. PubMed PMID: 26447401. | ||
| + | * Beumer KJ, Carroll D. [http://www.ncbi.nlm.nih.gov/pubmed/24412316 '''Targeted genome engineering techniques in Drosophila.'''] Methods. 2014 Jun 15;68(1):29-37. doi: 10.1016/j.ymeth.2013.12.002. Epub 2014 Jan 8. Review. PubMed PMID: 24412316; PubMed Central PMCID: PMC4048800. | ||
| + | * Xu J, Ren X, Sun J, Wang X, Qiao HH, Xu BW, Liu LP, Ni JQ. [http://www.ncbi.nlm.nih.gov/pubmed/25953352 '''A Toolkit of CRISPR-Based Genome Editing Systems in Drosophila. J Genet Genomics.'''] 2015 Apr 20;42(4):141-9. doi: 10.1016/j.jgg.2015.02.007. Epub 2015 Mar 12. Review. PubMed PMID: 25953352. | ||
| + | |||
| + | === Selected recent methods reports for tissue culture cells === | ||
| + | |||
| + | * Bassett AR, Tibbit C, Ponting CP, Liu JL. [http://www.ncbi.nlm.nih.gov/pubmed/24326186 '''Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9.'''] Biol Open. 2014 Jan 15;3(1):42-9. doi: 10.1242/bio.20137120. PMID: 24326186. | ||
| + | * Böttcher R, Hollmann M, Merk K, Nitschko V, Obermaier C, Philippou-Massier J, Wieland I, Gaul U, Förstemann K. [http://www.ncbi.nlm.nih.gov/pubmed/24748663 '''Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells.'''] Nucleic Acids Res. 2014 Jun;42(11):e89. doi: 10.1093/nar/gku289. Epub 2014 Apr 19. PMID: 24748663 | ||
| + | * Bassett AR, Kong L, Liu JL. [http://www.ncbi.nlm.nih.gov/pubmed/24748663 '''A genome-wide CRISPR library for high-throughput genetic screening in Drosophila cells.'''] J Genet Genomics. 2015 Jun 20;42(6):301-9. doi: 10.1016/j.jgg.2015.03.011. Epub 2015 Apr 18. PMID: 26165496 | ||
| + | * Housden BE, Valvezan AJ, Kelley C, Sopko R, Hu Y, Roesel C, Lin S, Buckner M, Tao R, Yilmazel B, Mohr SE, Manning BD, Perrimon N. [http://www.ncbi.nlm.nih.gov/pubmed/26350902 '''Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi.'''] Sci Signal. 2015 Sep 8;8(393):rs9. doi: 10.1126/scisignal.aab3729. PMID: 26350902 | ||
| + | |||
| + | === Selected recent methods reports for <em>in vivo</em> === | ||
| + | * Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. [http://www.ncbi.nlm.nih.gov/pubmed/23709638 '''Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease.'''] Genetics. 2013 Aug;194(4):1029-35. doi: 10.1534/genetics.113.152710. Epub 2013 May 24. PMID: 23709638 | ||
| + | |||
| + | * Bassett AR, Tibbit C, Ponting CP, Liu JL. [http://www.ncbi.nlm.nih.gov/pubmed/23827738 '''Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system.'''] Cell Rep. 2013 Jul 11;4(1):220-8. doi: 10.1016/j.celrep.2013.06.020. Epub 2013 Jul 1. PMID: 23827738 | ||
| + | |||
| + | * Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. [http://www.ncbi.nlm.nih.gov/pubmed/23833182 '''Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila.'''] Genetics. 2013 Sep;195(1):289-91. doi: 10.1534/genetics.113.153825. Epub 2013 Jul 5. PMID: 23833182 | ||
| + | |||
| + | * Kondo S, Ueda R. [http://www.ncbi.nlm.nih.gov/pubmed/24002648 '''Highly improved gene targeting by germline-specific Cas9 expression in Drosophila.'''] Genetics. 2013 Nov;195(3):715-21. doi: 10.1534/genetics.113.156737. Epub 2013 Sep 3. PMID: 24002648 | ||
| + | |||
| + | * Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu LP, Yang Z, Mao D, Sun L, Wu Q, Ji JY, Xi J, Mohr SE, Xu J, Perrimon N, Ni JQ. [http://www.ncbi.nlm.nih.gov/pubmed/24191015 '''Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9.'''] Proc Natl Acad Sci U S A. 2013 Nov 19;110(47):19012-7. doi: 10.1073/pnas.1318481110. Epub 2013 Nov 4. PMID: 24191015 | ||
| + | |||
| + | * Port F, Muschalik N, Bullock SL. [http://www.ncbi.nlm.nih.gov/pubmed/25999583 '''Systematic Evaluation of Drosophila CRISPR Tools Reveals Safe and Robust Alternatives to Autonomous Gene Drives in Basic Research.'''] G3 (Bethesda). 2015 May 20;5(7):1493-502. doi: 10.1534/g3.115.019083. PMID: 25999583 | ||
| + | |||
| + | * Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang SJ, Qiao HH, Wang X, Hu Q, Deng P, Liu LP, Ji JY, Li JB, Ni JQ. [http://www.ncbi.nlm.nih.gov/pubmed/25437567 '''Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila.'''] Cell Rep. 2014 Nov 6;9(3):1151-62. doi: 10.1016/j.celrep.2014.09.044. Epub 2014 Oct 23. PMID: 25437567 | ||
| + | <br> | ||
| + | [[Resource_Portal#top|Back to top.]] | [http://flybase.org/cgi-bin/mailto-fbhelp.html Provide Feedback.] | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | == RNAi == | ||
| + | |||
| + | |||
| + | === RNAi Design/Analysis Tools === | ||
| + | |||
| + | {|cellpadding=5 | ||
|- | |- | ||
| − | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source | |
|- | |- | ||
| − | | [http://www. | + | |style="background: #efefef; white-space"| [http://www.dkfz.de/signaling/e-rnai3/ E-RNAi]||style="background: #efefef; white-space:"| E-RNAi is a tool for the design and evaluation of RNAi reagents. It can be used to design and evaluate long dsRNAs (including esiRNAs) as well as siRNAs. ||style="background: #efefef;"| Boutros lab, DKFZ<br/> Heidelberg, Germany |
|- | |- | ||
| − | | | + | | [http://b110-wiki.dkfz.de/confluence/display/nextrnai/NEXT-RNAi NEXT-RNAi] || Software for the design and evaluation of genome-wide RNAi libraries which performs all steps from the prediction of specific and efficient RNAi target sites to the visualization of designed reagents in their genomic context. || Boutros lab, DFKZ<br/> Heidelberg, Germany |
|- | |- | ||
| − | | | + | |style="background: #efefef;"| [http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl SnapDragon] ||style="background: #efefef;"| A tool for the design of long dsRNAs for specific cell-based gene knockdown in Drosophila melanogaster via an RNAi approach. ||style="background: #efefef; white-space: nowrap"| Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA |
|} | |} | ||
| + | <br/> | ||
| − | === | + | === RNAi Stocks for in vivo studies (fly stocks) === |
{|cellpadding=5 | {|cellpadding=5 | ||
|- | |- | ||
| − | !style="background: #efefef;"| Resource !!style="background: #efefef;"| | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source |
|- | |- | ||
| − | + | | [http://www.flyrnai.org/up-torr/ UP-TORR, Updated Targets of RNAi Reagents] || A lookup tool for searching BDSC, TRiP, VDRC and NIG_FLY in vivo RNAi collections. || Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA | |
|- | |- | ||
| − | | [http:// | + | |style="background: #efefef; white-space:"| [http://flystocks.bio.indiana.edu/Browse/RNAi/RNAihome.htm BDSC RNAi Home Page]||style="background: #efefef; white-space:"| Links to lists and sublists of RNAi and miRNA sponge insertions at Bloomington. Most, but not all, of the insertions are under the control of UAS and therefore require GAL4 for expression. ||style="background: #efefef; white-space:nowrap"| Bloomington Drosophila Stock Center (BDSC)<br/> Indiana University<br/> Bloomington, IN, USA |
|- | |- | ||
| − | | | + | | [http://www.shigen.nig.ac.jp/fly/nigfly/index.jsp NIG_FLY] || NIG-Fly in Mishima, Japan is part of a consortium with Kyoto-Fly and two other stock centers, and distributes RNAi flies on request. || NIG-FLY<br/> Mishima, Japan |
|- | |- | ||
| − | | | + | |style="background: #efefef;"| [http://www.flyrnai.org/TRiP-HOME.html TRiP Transgenic RNAi Project] ||style="background: #efefef;"| The TRiP Stock Collection contains over 9,000 lines, 1,575 of which are for orthologs of human-disease-associated genes. The stocks can be obtained from the BDSC. ||style="background: #efefef;"| Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA |
| + | |- | ||
| + | | [http://center.biomed.tsinghua.edu.cn/public/eq-category/modelanimalfacility/ THFC, Tsinghua Fly Center] || THFC RNAi Stock Collection for triggering RNAi in soma and germline || Tsinghua Fly Center<br/> Beijing, China | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://stockcenter.vdrc.at/control/main VDRC, Vienna Drosophila Resource Center] ||style="background: #efefef;"| The VDRC at IMP/IMBA in Vienna provides two genome-wide transgenic Drosophila RNAi libraries ||style="background: #efefef;"| VDRC<br/> Vienna, Austria | ||
|} | |} | ||
| + | <br/> | ||
| − | === | + | === Cell-based RNAi Reagents === |
{|cellpadding=5 | {|cellpadding=5 | ||
|- | |- | ||
| − | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Author !!style="background: #efefef;"| | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source |
| + | |- | ||
| + | |style="background: #efefef; white-space:"| [http://www.flyrnai.org/DRSC-DRS.html DRSC Libraries]||style="background: #efefef; white-space:"| Genome-Wide Libraries for cell-based RNAi Screening and Validation (on-site or shipped) ||style="background: #efefef;"| Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA | ||
| + | |- | ||
| + | | [http://www.dkfz.de/signaling/ Division of Signaling and Functional Genomics] || Genome-Wide Libraries for cell-based RNAi Screening and Validation || Boutros lab, DKFZ, Heidelberg, Germany | ||
| + | |} | ||
| + | <br/> | ||
| + | |||
| + | === RNAi Screening Centers === | ||
| + | |||
| + | {|cellpadding=5 | ||
| + | |- | ||
| + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source | ||
| + | |- | ||
| + | |style="background: #efefef; white-space"| [http://www.flyrnai.org/RNAi_index.html DRSC, Drosophila RNAi Screening Center]||style="background: #efefef; white-space:"| The Drosophila RNAi Screening Center at Harvard Medical School provides high throughput RNAi screens. ||style="background: #efefef;"| Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA | ||
| + | |- | ||
| + | |style="nowrap;"| [http://www.med.nyu.edu/ocs/rnai/ NYU RNAi Screening and Automation Core] || The NYU RNAi Core Facility at the Langone Medical Center in New York provides high-throughput RNAi screens. Drosophila screens are offered in collaboration with the DRSC. || NYU RNAi Core, New York, NY, USA | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.rnai.group.shef.ac.uk/ Sheffield RNAi Screening Facility] ||style="background: #efefef;"| The SRSF provides a service for whole-genome RNAi screens in Drosophila cells. ||style="background: #efefef;"| SRSF, University of Sheffield, Boston, MA, USA | ||
| + | |} | ||
| + | <br/> | ||
| + | |||
| + | === RNAi Results, Validation and Phenotypes === | ||
| + | |||
| + | {|cellpadding=5 | ||
|- | |- | ||
| − | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source | |
|- | |- | ||
| − | | [http://www. | + | |style="background: #efefef; white-space"| [http://www.genomernai.org/ GenomeRNAi] ||style="background: #efefef; white-space:"| GenomeRNAi is a database containing phenotypes from cell-based and in vivo RNA interference (RNAi) screens in Drosophila. ||style="background: #efefef;"| GenomeRNAi, DKFZ, Heidelberg, Germany |
|- | |- | ||
| − | |style=" | + | |style="nowrap;"| [http://www.flyrnai.org/RSVP RSVP] || RNAi Stock Validation and Phenotypes for in vivo RNAi studies || Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA |
|- | |- | ||
| − | | | + | |style="background: #efefef;"| [http://www.flyrnai.org/screensummary Cell-Based RNAi Screen Summary] ||style="background: #efefef;"| A summary of public cell-based DRSC screens ||style="background: #efefef;"| Drosophila RNAi Screening Center (DRSC)<br/> Harvard Medical School<br/> Boston, MA, USA |
|} | |} | ||
| − | + | <br> | |
| + | [[Resource_Portal#top|Back to top.]] | [http://flybase.org/cgi-bin/mailto-fbhelp.html Provide Feedback.] | ||
| + | |||
| + | <br><br> | ||
| − | == | + | ==Stock Centers== |
| − | |||
| − | === | + | === List of D. melanogaster Stock Collections === |
{|cellpadding=5 | {|cellpadding=5 | ||
|- | |- | ||
| − | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Author !!style="background: #efefef;"| | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source |
| + | |- | ||
| + | |style="background: #efefef;"| [http://flystocks.bio.indiana.edu/ BDSC, Bloomington Drosophila Stock Center] ||style="background: #efefef;"| A diverse collection of stocks useful for a wide range of research applications. ||style="background: #efefef;"| BDSC, Indiana University, Bloomington, IN, USA | ||
| + | |- | ||
| + | | [https://kyotofly.kit.jp/cgi-bin/stocks/index.cgi DGGR, Kyoto Stock Center] || The Department of Drosophila Genomics and Genetics Resources collects, maintains and distributes Drosophila melanogaster strains for research. || DGGR, Kyoto, Japan | ||
| + | |- | ||
| + | |style="background: #efefef;"| [https://drosophila.med.harvard.edu/ Exelixis] ||style="background: #efefef;"| The Exelixis Collection at Harvard Medical School||style="background: #efefef;"| Harvard Medical School, Boston, MA, USA | ||
| + | |- | ||
| + | | [http://flyorf.ch/ FlyORF] || Well characterized transgenic UAS-ORF lines. Approximately 2,400 fly stocks comprising about 1900 genes, generated using the phiC31 integrase method || FlyORF, University of Zurich, Zurich, Switzerland | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.shigen.nig.ac.jp/fly/nigfly/ NIG-FLY] ||style="background: #efefef;"| NIG-RNAi stocks and TRiP Stocks for RNA interference experiments ||style="background: #efefef;"| NIG-FLY, National Institute of Genetics, Mishima, Japan | ||
| + | |- | ||
| + | |style="white-space: nowrap;"| [http://flystocks.bio.indiana.edu/Browse/misc-browse/CRISPR.htm CRISPR system stocks for site-specific mutagenesis]||style=" white-space: nowrap;"| CRISPR system stocks for site-specific mutagenesis at BDSC || BDSC, Indiana University, Bloomington, IN, USA | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://center.biomed.tsinghua.edu.cn/public/eq-category/modelanimalfacility/ Tsinghua Fly Center (THFC)] ||style="background: #efefef;"| Transgenic RNAi fly lines ||style="background: #efefef;"| THFC, Tsinghua University, Beijing, China | ||
| + | |- | ||
| + | | [http://stockcenter.vdrc.at/control/main VDRC, Vienna Drosophila RNAi Center] || Trangenic Drosophila RNAi libraries, a collection of enhancer-GAL4 driver lines, the Tagged FlyFos TransgeneOme (fTRG) library || VDRC, Vienna, Austria | ||
| + | |} | ||
| + | <br/> | ||
| + | |||
| + | === D. melanogaster Stock Collection Informational Sites === | ||
| + | |||
| + | {|cellpadding=5 | ||
| + | |- | ||
| + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.crisprflydesign.org/flies/ CRISPR fly design] ||style="background: #efefef;"| Details of a set of cas9 transgenic D. melanogaster lines which are available at the BDSC ||style="background: #efefef;"| MRC-Laboratory of Molecular Biology, Cambridge, UK | ||
| + | |- | ||
| + | | [http://www.drosdel.org.uk/ DrosDel An isogenic deficiency kit for Drosophila melanogaster]|| Information about isogenic deficiency lines generated by the DrosDel Project || University of Cambridge, Cambridge, UK | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://cooley.medicine.yale.edu/flytrap/ Flytrap Project] ||style="background: #efefef;"| Information about GFP protein trap lines from the FlyTrap project ||style="background: #efefef;"| Yale University, New Haven, CT, USA | ||
| + | |- | ||
| + | | [http://flypush.imgen.bcm.tmc.edu/pscreen/ Gene Disruption Project] || Information about insertion lines produced by the Gene Disruption Project (GDP) || Baylor College of Medicine, Houston, TX, USA | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://flypush.imgen.bcm.tmc.edu/pscreen/rmce/ Gene Disruption Project] ||style="background: #efefef;"| Information about MiMIC RMCE Lines ||style="background: #efefef;"| Baylor College of Medicine, Houston, TX, USA | ||
| + | |- | ||
| + | | [http://www.ehime-u.ac.jp/~scibio/evolut/DSEU_photo/ Ehime University fly species photographs] || Photograph of 102 species of Drosophila in Drosophila stocks of Ehime University. || Ehime University, Matsuyama, Ehime, Japan | ||
| + | |} | ||
| + | <br/> | ||
| + | |||
| + | === List of Drosophila Species Collections === | ||
| + | |||
| + | {|cellpadding=5 | ||
|- | |- | ||
| − | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Description !!style="background: #efefef;"| Author/Source | |
|- | |- | ||
| − | | [ | + | |style="background: #efefef; white-space: nowrap;"| [https://kyotofly.kit.ac.jp/cgi-bin/ehime/index.cgi EHIME-Fly]||style="background: #efefef;"| Wild type stocks of Drosophila species that have been recorded in Japan. EHIME-Fly maintains and distributes over 130 species, represented by approximately 1450 stocks. ||style="background: #efefef; white-space: nowrap;"| Ehime University<br/> Matsuyama, Ehime Prefecture, Japan |
|- | |- | ||
| − | | | + | | [https://stockcenter.ucsd.edu/info/welcome.php DSSC, Drosophila Species Stock Center] || The DSSC maintains a living collection of over 250 Drosophila species represented by approximately 1600 stocks || University of California San Diego<br/> San Diego, CA, USA |
|- | |- | ||
| − | | | + | |style="background: #efefef; white-space: nowrap;"| [http://www.shigen.nig.ac.jp/fly/kyorin/cgi-bin/index.cgi KYORIN-Fly ] ||style="background: #efefef;"| Mutant and wild type stocks of Drosophila species, especially the Drosophila ananassae species subgroup, D. auraria complex, and D. hydei. ||style="background: #efefef;"| Kyorin University<br/> Shinkawa, Mitaka, Tokyo, Japan |
|} | |} | ||
| − | === | + | |
| + | <br> | ||
| + | [[Resource_Portal#top|Back to top.]] | [http://flybase.org/cgi-bin/mailto-fbhelp.html Provide Feedback.] | ||
| + | |||
| + | == Antibodies == | ||
| + | |||
| + | ===Links to collections of <em>Drosophila</em>-specific antibodies.=== | ||
{|cellpadding=5 | {|cellpadding=5 | ||
|- | |- | ||
| − | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Author | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Author/Source |
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.abbiotec.com/antibodies?populate=&field_antibody_subcategory_tid=All&field_isotype_value=All&field_species_reactivity_value=Ds&field_applications_value=All&field_protein_family_tid=All Abbiotec] ||style="background: #efefef;"|San Diego, CA, USA | ||
| + | |- | ||
| + | |[http://www.abcam.com/products?keywords=drosophila&selected.classification=Primary+antibodies&selected.reactivity=Fruit+fly+(Drosophila+melanogaster) Abcam] ||Cambridge, MA, USA | ||
| + | |- | ||
| + | |style="background: #efefef;"|[https://us.acris-antibodies.com/antibodies/primary-antibodies.htm?ab_ag_reactivity=Dros Acris Antibodies] ||style="background: #efefef;"|San Diego, CA, USA<br/> Herford, Germany | ||
| + | |- | ||
| + | |[https://www.activemotif.com/catalog/806/search-browse-all-antibodies?svs_product_class=Antibodies&mvs_reactivity=Drosophila Active Motif] ||Carlsbad, CA, USA <br/>International Locations | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.biorbyt.com/products/antibodies?speciesselect=51 Biorbyt] ||style="background: #efefef;"| Cambridge, UK<br/> | ||
|- | |- | ||
| − | + | | [http://www.cellsignal.com/browse/d-melanogaster?N=4294956287+4294964832+4294960387 Cell Signaling Technology] || Danvers, MA, USA<br/> International Locations | |
|- | |- | ||
| − | | [http:// | + | |style="background: #efefef;"| [http://dshb.biology.uiowa.edu/Antibody-Collections/Drosophila-antigens Developmental Studies Hybridoma Bank (DSHB)] ||style="background: #efefef;"| University of Iowa<br/> |
| + | Iowa City, IA, USA | ||
|- | |- | ||
| − | | | + | | [http://fabgennix.com/epages/862cc93a-7f43-4d98-8516-1e11a7720fd8.sf/en_US/?ObjectID=81707&ViewAction=ViewFaceted&FacetValue_CategoryID=81707&FacetValue_PreDefString_en_1540487778=D.+melanogaster&CurrencyID=USD&CurrencyID=USD&FacetRange_ListPrice=&FacetRange_ListPrice= FabGennix] || Frisco, TX, USA |
|- | |- | ||
| − | | | + | |style="background: #efefef;"| [http://www.genetex.com/Web/search/Catalog.aspx?Type=1&Category=494&Reactivity=Drosophila&Page=1 GeneTex] ||style="background: #efefef;"| Irvine, CA, USA<br/>International Locations |
| + | |- | ||
| + | || [http://www.novusbio.com/drosophilaantibodies.html Novus]|| Littleton, CO, USA | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.prosci-inc.com/primary-antibodies.html?species_reactivity=601 ProSci] ||style="background: #efefef;"| Poway, CA, USA | ||
| + | |- | ||
| + | || [https://www.rndsystems.com/search?keywords=primary%20antibodies&species=Drosophila R&D Systems]|| Minneapolis, MN, USA | ||
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.scbt.com/section/primary_antibodies_non-mammalian/drosophila_proteins.html Santa Cruz Biotechnology]||style="background: #efefef;"| Dallas, Texas, USA | ||
| + | |- | ||
| + | || [http://www.sigmaaldrich.com/catalog/search?interface=All&N=0+14574647+4294666762&mode=partialmax&focus=product&lang=en®ion=US Sigma Aldrich] ([http://www.sigmaaldrich.com/catalog/search?interface=All&N=0+14574647+4294666151&mode=partialmax&focus=product&lang=en®ion=US additional link])|| St. Louis, MO, USA <br/> International Locations | ||
|} | |} | ||
| + | <br> | ||
| − | == | + | ===Links to vendors whose collections include <em>Drosophila</em>-specific antibodies.=== |
| − | + | The table below contains non-direct <em>Drosophila</em> antibody links, additional searching is required from the vendor website. | |
{|cellpadding=5 | {|cellpadding=5 | ||
|- | |- | ||
| − | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Author | + | !style="background: #efefef;"| Resource !!style="background: #efefef;"| Author/Source |
| + | |- | ||
| + | |style="background: #efefef;"| [http://www.abgent.com/ Abgent] ||style="background: #efefef; whitespace:no-wrap;"|San Diego, CA, USA <br/> Suzhou city, Jiangsu Province, China | ||
|- | |- | ||
| − | + | |[http://www.antibodies-online.com/ antibodies-online] ||International Locations | |
|- | |- | ||
| − | | [http://www. | + | |style="background: #efefef;"|[http://www.enzolifesciences.com/ Enzo Life Sciences] ||style="background: #efefef;"|Farmingdale, NY, USA |
|- | |- | ||
| − | | | + | |[https://www.lsbio.com/ LifeSpan BioSciences] ||Seattle, WA, USA |
|- | |- | ||
| − | | | + | |style="background: #efefef;"|[https://www.thermofisher.com/us/en/home.html Thermo Fisher] || style="background: #efefef;"|Waltham, MA, USA |
|} | |} | ||
| + | |||
| + | <br/> | ||
| + | |||
| + | ===Lab-generated antibodies in Gene Reports=== | ||
| + | |||
| + | Antibodies generated by research laboratories for specific proteins are listed in their respective Gene Report under "'''Stocks and Reagents'''" → "'''Antibody Information'''". | ||
| + | <br/> | ||
| + | For example, the following screenshot is taken from the [http://flybase.org/reports/FBgn0004647.html Notch gene report]:<br/> <br/> | ||
| + | [[File:Antibodies.png|800px]] | ||
| + | <br/><br/> | ||
| + | If no antibodies exist or they have not yet been curated, this section will be empty. | ||
| + | |||
| + | <br> | ||
| + | [[Resource_Portal#top|Back to top.]] | [http://flybase.org/cgi-bin/mailto-fbhelp.html Provide Feedback.] | ||
| + | |||
| + | =All Resources= | ||
| + | An extensive list of resources and reagents can be found on the pages linked below: | ||
| + | |||
| + | <big>'''[[FlyBase:Drosophila Network Resources|Drosophila Network Resources]]'''</big> | ||
| + | |||
| + | Includes: | ||
| + | *[[FlyBase:Drosophila_Network_Resources#Atlases.2C_Images_and_Videos|Atlases, Images, and Videos]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#CRISPRs_and_TALENs|CRISPRs and TALENs]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Data_Repositories|Data Repositories]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Data_and_Metadata_for_Drosophila_Genomes|Data and Metadata for Drosophila Genomes]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Gene_Expression_Databases_and_Tools|Gene Expression Databases and Tools]] | ||
| + | *[[FlyBase:Drosophila_Network_Resources#Gene_Groups|Gene Groups]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#General_Bioinformatics_Tools|General Bioinformatics Tools]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Genome_Sequencing_Projects|Genome Sequencing Projects]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Human_Disease:_Drosophila_Models_and_Orthologous_Genes|Human Disease: Drosophila Models and Orthologous Genes]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Interaction_and_Pathway_Databases|Interaction and Pathway Databases]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Laboratory_Resources|Laboratory Resources]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#miRNA_and_ncRNA_Databases_and_Tools|miRNA and ncRNA Databases and Tools]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Miscellaneous|Miscellaneous]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Ontology_Resources|Ontology Resources]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Ontology_Resources|Orthologs]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Phylogenetic_Comparison_Tools|Phylogenetic Comparison Tools]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Population_Biology_and_Polymorphism_Resources|Population Biology and Polymorphism Resources]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Protein_Analysis|Protein Analysis]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Public_Education|Public Education]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#RNAi|RNAi]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Sequence_Analysis|Sequence Analysis]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Taxonomy|Taxonomy]] | ||
| + | * [[FlyBase:Drosophila_Network_Resources#Transcription_Regulation_Databases_and_Tools|Transcription Regulation Databases and Tools]] | ||
| + | |||
| + | <big>'''[[FlyBase:Drosophila Material Resources|Drosophila Material Resources (Reagents)]]'''</big> | ||
| + | |||
| + | Includes: | ||
| + | * [[FlyBase:Drosophila_Material_Resources#Genomic_Clones|Genomic Clones]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#cDNA_.28EST_Clones_and_Libraries.29|cDNA (EST Clones and Libraries)]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#Arrays_and_Primers|Arrays and Primers]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#Stocks|Stocks]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#Stocks|CRISPR Vectors]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#Expression_Vectors|Expression Vectors]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#Cell_Lines_and_Hybridomas|Cell Lines and Hybridomas]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#Antibodies|Antibodies]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#Transgenic.2C_Targeted_Mutation.2C_and_Other_Services|Transgenic, Targeted Mutation, and Other Services]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#RNA|RNA]] | ||
| + | * [[FlyBase:Drosophila_Material_Resources#RNAi|RNAi]] | ||
| + | |||
| + | <br> | ||
| + | [[Resource_Portal#top|Back to top.]] | [http://flybase.org/cgi-bin/mailto-fbhelp.html Provide Feedback.] | ||
Latest revision as of 20:17, 9 December 2015
FlyBase maintains extensive lists of useful external Network and Material Resources for fly researchers that can be accessed from the following links:
Network Resources
Reagent Resources
Links relevant to some of our most popular resources have been compiled and are listed below.
Popular Resources
| CRISPR | RNAi | Stock Centers | Antibodies | Orthologs | All Resources |
Full Table of Contents
CRISPR
gRNA Design Resources
| Resource | Description | Author/Source |
|---|---|---|
| CRISPR Optimal Target Finder | Identifies gRNA targets within a provided sequence and additionally searches genome-wide (release 6, current FlyBase release) for potential off-target sites. Includes numerous Drosophila species. | O'Connor-Giles, Wildonger, and Harrison Labs University of Wisconsin-Madison WI, USA |
| CRISPRscan | Searchable and browsable collection of genome-wide (release 6, current FlyBase release) sgRNA target sites. Displays sgRNA sites as tracks on UCSC's genome browser and allows searching via gene or prediction via sequence. Support for numerous species (fly, zebrafish, mouse, human, others). | Giraldez Lab Yale University New Haven, CT, USA |
| DRSC Find CRISPR Tool | A tool for discovering gRNA targets located throughout the fly genome (release 5) using gene IDs, symbols, or chromosome locations. Allows for filtering of off-target sites and control of mis-match stringency. | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
| E-CRISPR | gRNA genome-wide (release 5) target finder, searchable via gene symbol or sequence. Support for numerous species (fly, zebrafish, mouse, human, others). Offers "relaxed, medium and strict" search options. | Boutros lab, DKFZ Heidelberg, Germany |
| CHOPCHOP | Search for gRNA targets (release 5). Flexible inputs (gene name, genomic coordinates or DNA sequence) and detection of off-target sites. | Schier & Church Labs Harvard University Cambridge, MA, USA |
| CRISPR Design | Identification of gRNA sites genome wide (release 5). Offers options for multiple species. | Zhang Lab MIT Boston, MA, USA |
| CRISPOR | A program that helps design, evaluate, and clone guide sequences for the CRISPR/Cas9 system. Allows for searches across multiple species of Drosophila, including melanogaster (release 5). | Tefor France |
CRISPR Stocks
| Resource | Description | Author/Source |
|---|---|---|
| CRISPR system stocks for site-specific mutagenesis | The Bloomington Stock Center's CRISPR/Cas9 page. Includes various stocks for expression of both Cas9 and tracrRNA. | Bloomington Drosophila Stock Center (BDSC) Indiana University Bloomington, IN, USA |
| CRISPR Fly Design | Information on stocks generated by the CRISPR Fly Design group at the MRC Laboratory of Molecular Biology. | MRC Laboratory of Molecular Biology Cambridge, UK |
CRISPR Vectors
| Resource | Description | Author/Source |
|---|---|---|
| Addgene CRISPR/Cas9 Plasmids for use in insects | A list of available Cas9 & gRNA expression plasmids for use in insects, including Drosophila. | Addgene Cambridge, MA, USA |
| CRISPR Fly Design Plasmids | Descriptions and information regarding the plasmids created by the CRISPR Fly Design group (also deposited at Addgene). | MRC Laboratory of Molecular Biology Cambridge, UK |
Additional Useful Links
| Resource | Description | Author/Source |
|---|---|---|
| CRISPR Fly Design | An overview and resource collection for CRISPR gene editing in Drosophila. | MRC Laboratory of Molecular Biology Cambridge, UK |
| flyCRISPR | An overview and resource collection for CRISPR gene editing in Drosophila. | O'Connor-Giles, Wildonger, and Harrison Labs University of Wisconsin-Madison WI, USA |
| Insect Genomic Technologies Research Coordination Network | Discussion and dissemination of advanced technologies for genome modification of insects through symposia, technical workshops, and training fellowships. | Institute for Bioscience and Biotechnology Research University of Maryland Rockville, MD, USA |
| FlyRNAi Blog, CRISPR tags | A research blog tracking functional genomics techniques in Drosophila, filtered for keyword: CRISPRs. | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
| DRSC Single Cell Cloning Protocol | Single-cell cloning method useful for Drosophila cell CRISPR gene editing | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
Selected recent methods reviews
- Venken KJ, Sarrion-Perdigones A, Vandeventer PJ, Abel NS, Christiansen AE, Hoffman KL. Genome engineering: Drosophila melanogaster and beyond. Wiley Interdiscip Rev Dev Biol. 2015 Oct 8. doi: 10.1002/wdev.214. [Epub ahead of print] Review. PubMed PMID: 26447401.
- Beumer KJ, Carroll D. Targeted genome engineering techniques in Drosophila. Methods. 2014 Jun 15;68(1):29-37. doi: 10.1016/j.ymeth.2013.12.002. Epub 2014 Jan 8. Review. PubMed PMID: 24412316; PubMed Central PMCID: PMC4048800.
- Xu J, Ren X, Sun J, Wang X, Qiao HH, Xu BW, Liu LP, Ni JQ. A Toolkit of CRISPR-Based Genome Editing Systems in Drosophila. J Genet Genomics. 2015 Apr 20;42(4):141-9. doi: 10.1016/j.jgg.2015.02.007. Epub 2015 Mar 12. Review. PubMed PMID: 25953352.
Selected recent methods reports for tissue culture cells
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol Open. 2014 Jan 15;3(1):42-9. doi: 10.1242/bio.20137120. PMID: 24326186.
- Böttcher R, Hollmann M, Merk K, Nitschko V, Obermaier C, Philippou-Massier J, Wieland I, Gaul U, Förstemann K. Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res. 2014 Jun;42(11):e89. doi: 10.1093/nar/gku289. Epub 2014 Apr 19. PMID: 24748663
- Bassett AR, Kong L, Liu JL. A genome-wide CRISPR library for high-throughput genetic screening in Drosophila cells. J Genet Genomics. 2015 Jun 20;42(6):301-9. doi: 10.1016/j.jgg.2015.03.011. Epub 2015 Apr 18. PMID: 26165496
- Housden BE, Valvezan AJ, Kelley C, Sopko R, Hu Y, Roesel C, Lin S, Buckner M, Tao R, Yilmazel B, Mohr SE, Manning BD, Perrimon N. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci Signal. 2015 Sep 8;8(393):rs9. doi: 10.1126/scisignal.aab3729. PMID: 26350902
Selected recent methods reports for in vivo
- Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O'Connor-Giles KM. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013 Aug;194(4):1029-35. doi: 10.1534/genetics.113.152710. Epub 2013 May 24. PMID: 23709638
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013 Jul 11;4(1):220-8. doi: 10.1016/j.celrep.2013.06.020. Epub 2013 Jul 1. PMID: 23827738
- Yu Z, Ren M, Wang Z, Zhang B, Rong YS, Jiao R, Gao G. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 2013 Sep;195(1):289-91. doi: 10.1534/genetics.113.153825. Epub 2013 Jul 5. PMID: 23833182
- Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013 Nov;195(3):715-21. doi: 10.1534/genetics.113.156737. Epub 2013 Sep 3. PMID: 24002648
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, Liu LP, Yang Z, Mao D, Sun L, Wu Q, Ji JY, Xi J, Mohr SE, Xu J, Perrimon N, Ni JQ. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci U S A. 2013 Nov 19;110(47):19012-7. doi: 10.1073/pnas.1318481110. Epub 2013 Nov 4. PMID: 24191015
- Port F, Muschalik N, Bullock SL. Systematic Evaluation of Drosophila CRISPR Tools Reveals Safe and Robust Alternatives to Autonomous Gene Drives in Basic Research. G3 (Bethesda). 2015 May 20;5(7):1493-502. doi: 10.1534/g3.115.019083. PMID: 25999583
- Ren X, Yang Z, Xu J, Sun J, Mao D, Hu Y, Yang SJ, Qiao HH, Wang X, Hu Q, Deng P, Liu LP, Ji JY, Li JB, Ni JQ. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014 Nov 6;9(3):1151-62. doi: 10.1016/j.celrep.2014.09.044. Epub 2014 Oct 23. PMID: 25437567
Back to top. | Provide Feedback.
RNAi
RNAi Design/Analysis Tools
| Resource | Description | Author/Source |
|---|---|---|
| E-RNAi | E-RNAi is a tool for the design and evaluation of RNAi reagents. It can be used to design and evaluate long dsRNAs (including esiRNAs) as well as siRNAs. | Boutros lab, DKFZ Heidelberg, Germany |
| NEXT-RNAi | Software for the design and evaluation of genome-wide RNAi libraries which performs all steps from the prediction of specific and efficient RNAi target sites to the visualization of designed reagents in their genomic context. | Boutros lab, DFKZ Heidelberg, Germany |
| SnapDragon | A tool for the design of long dsRNAs for specific cell-based gene knockdown in Drosophila melanogaster via an RNAi approach. | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
RNAi Stocks for in vivo studies (fly stocks)
| Resource | Description | Author/Source |
|---|---|---|
| UP-TORR, Updated Targets of RNAi Reagents | A lookup tool for searching BDSC, TRiP, VDRC and NIG_FLY in vivo RNAi collections. | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
| BDSC RNAi Home Page | Links to lists and sublists of RNAi and miRNA sponge insertions at Bloomington. Most, but not all, of the insertions are under the control of UAS and therefore require GAL4 for expression. | Bloomington Drosophila Stock Center (BDSC) Indiana University Bloomington, IN, USA |
| NIG_FLY | NIG-Fly in Mishima, Japan is part of a consortium with Kyoto-Fly and two other stock centers, and distributes RNAi flies on request. | NIG-FLY Mishima, Japan |
| TRiP Transgenic RNAi Project | The TRiP Stock Collection contains over 9,000 lines, 1,575 of which are for orthologs of human-disease-associated genes. The stocks can be obtained from the BDSC. | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
| THFC, Tsinghua Fly Center | THFC RNAi Stock Collection for triggering RNAi in soma and germline | Tsinghua Fly Center Beijing, China |
| VDRC, Vienna Drosophila Resource Center | The VDRC at IMP/IMBA in Vienna provides two genome-wide transgenic Drosophila RNAi libraries | VDRC Vienna, Austria |
Cell-based RNAi Reagents
| Resource | Description | Author/Source |
|---|---|---|
| DRSC Libraries | Genome-Wide Libraries for cell-based RNAi Screening and Validation (on-site or shipped) | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
| Division of Signaling and Functional Genomics | Genome-Wide Libraries for cell-based RNAi Screening and Validation | Boutros lab, DKFZ, Heidelberg, Germany |
RNAi Screening Centers
| Resource | Description | Author/Source |
|---|---|---|
| DRSC, Drosophila RNAi Screening Center | The Drosophila RNAi Screening Center at Harvard Medical School provides high throughput RNAi screens. | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
| NYU RNAi Screening and Automation Core | The NYU RNAi Core Facility at the Langone Medical Center in New York provides high-throughput RNAi screens. Drosophila screens are offered in collaboration with the DRSC. | NYU RNAi Core, New York, NY, USA |
| Sheffield RNAi Screening Facility | The SRSF provides a service for whole-genome RNAi screens in Drosophila cells. | SRSF, University of Sheffield, Boston, MA, USA |
RNAi Results, Validation and Phenotypes
| Resource | Description | Author/Source |
|---|---|---|
| GenomeRNAi | GenomeRNAi is a database containing phenotypes from cell-based and in vivo RNA interference (RNAi) screens in Drosophila. | GenomeRNAi, DKFZ, Heidelberg, Germany |
| RSVP | RNAi Stock Validation and Phenotypes for in vivo RNAi studies | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
| Cell-Based RNAi Screen Summary | A summary of public cell-based DRSC screens | Drosophila RNAi Screening Center (DRSC) Harvard Medical School Boston, MA, USA |
Back to top. | Provide Feedback.
Stock Centers
List of D. melanogaster Stock Collections
| Resource | Description | Author/Source |
|---|---|---|
| BDSC, Bloomington Drosophila Stock Center | A diverse collection of stocks useful for a wide range of research applications. | BDSC, Indiana University, Bloomington, IN, USA |
| DGGR, Kyoto Stock Center | The Department of Drosophila Genomics and Genetics Resources collects, maintains and distributes Drosophila melanogaster strains for research. | DGGR, Kyoto, Japan |
| Exelixis | The Exelixis Collection at Harvard Medical School | Harvard Medical School, Boston, MA, USA |
| FlyORF | Well characterized transgenic UAS-ORF lines. Approximately 2,400 fly stocks comprising about 1900 genes, generated using the phiC31 integrase method | FlyORF, University of Zurich, Zurich, Switzerland |
| NIG-FLY | NIG-RNAi stocks and TRiP Stocks for RNA interference experiments | NIG-FLY, National Institute of Genetics, Mishima, Japan |
| CRISPR system stocks for site-specific mutagenesis | CRISPR system stocks for site-specific mutagenesis at BDSC | BDSC, Indiana University, Bloomington, IN, USA |
| Tsinghua Fly Center (THFC) | Transgenic RNAi fly lines | THFC, Tsinghua University, Beijing, China |
| VDRC, Vienna Drosophila RNAi Center | Trangenic Drosophila RNAi libraries, a collection of enhancer-GAL4 driver lines, the Tagged FlyFos TransgeneOme (fTRG) library | VDRC, Vienna, Austria |
D. melanogaster Stock Collection Informational Sites
| Resource | Description | Author/Source |
|---|---|---|
| CRISPR fly design | Details of a set of cas9 transgenic D. melanogaster lines which are available at the BDSC | MRC-Laboratory of Molecular Biology, Cambridge, UK |
| DrosDel An isogenic deficiency kit for Drosophila melanogaster | Information about isogenic deficiency lines generated by the DrosDel Project | University of Cambridge, Cambridge, UK |
| Flytrap Project | Information about GFP protein trap lines from the FlyTrap project | Yale University, New Haven, CT, USA |
| Gene Disruption Project | Information about insertion lines produced by the Gene Disruption Project (GDP) | Baylor College of Medicine, Houston, TX, USA |
| Gene Disruption Project | Information about MiMIC RMCE Lines | Baylor College of Medicine, Houston, TX, USA |
| Ehime University fly species photographs | Photograph of 102 species of Drosophila in Drosophila stocks of Ehime University. | Ehime University, Matsuyama, Ehime, Japan |
List of Drosophila Species Collections
| Resource | Description | Author/Source |
|---|---|---|
| EHIME-Fly | Wild type stocks of Drosophila species that have been recorded in Japan. EHIME-Fly maintains and distributes over 130 species, represented by approximately 1450 stocks. | Ehime University Matsuyama, Ehime Prefecture, Japan |
| DSSC, Drosophila Species Stock Center | The DSSC maintains a living collection of over 250 Drosophila species represented by approximately 1600 stocks | University of California San Diego San Diego, CA, USA |
| KYORIN-Fly | Mutant and wild type stocks of Drosophila species, especially the Drosophila ananassae species subgroup, D. auraria complex, and D. hydei. | Kyorin University Shinkawa, Mitaka, Tokyo, Japan |
Back to top. | Provide Feedback.
Antibodies
Links to collections of Drosophila-specific antibodies.
| Resource | Author/Source |
|---|---|
| Abbiotec | San Diego, CA, USA |
| Abcam | Cambridge, MA, USA |
| Acris Antibodies | San Diego, CA, USA Herford, Germany |
| Active Motif | Carlsbad, CA, USA International Locations |
| Biorbyt | Cambridge, UK |
| Cell Signaling Technology | Danvers, MA, USA International Locations |
| Developmental Studies Hybridoma Bank (DSHB) | University of Iowa Iowa City, IA, USA |
| FabGennix | Frisco, TX, USA |
| GeneTex | Irvine, CA, USA International Locations |
| Novus | Littleton, CO, USA |
| ProSci | Poway, CA, USA |
| R&D Systems | Minneapolis, MN, USA |
| Santa Cruz Biotechnology | Dallas, Texas, USA |
| Sigma Aldrich (additional link) | St. Louis, MO, USA International Locations |
Links to vendors whose collections include Drosophila-specific antibodies.
The table below contains non-direct Drosophila antibody links, additional searching is required from the vendor website.
| Resource | Author/Source |
|---|---|
| Abgent | San Diego, CA, USA Suzhou city, Jiangsu Province, China |
| antibodies-online | International Locations |
| Enzo Life Sciences | Farmingdale, NY, USA |
| LifeSpan BioSciences | Seattle, WA, USA |
| Thermo Fisher | Waltham, MA, USA |
Lab-generated antibodies in Gene Reports
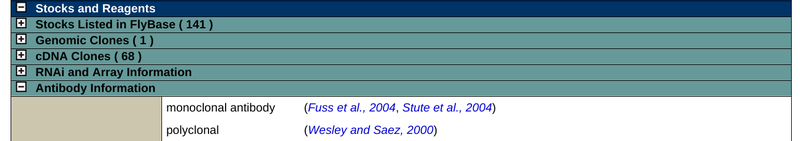
Antibodies generated by research laboratories for specific proteins are listed in their respective Gene Report under "Stocks and Reagents" → "Antibody Information".
For example, the following screenshot is taken from the Notch gene report:
If no antibodies exist or they have not yet been curated, this section will be empty.
Back to top. | Provide Feedback.
All Resources
An extensive list of resources and reagents can be found on the pages linked below:
Includes:
- Atlases, Images, and Videos
- CRISPRs and TALENs
- Data Repositories
- Data and Metadata for Drosophila Genomes
- Gene Expression Databases and Tools
- Gene Groups
- General Bioinformatics Tools
- Genome Sequencing Projects
- Human Disease: Drosophila Models and Orthologous Genes
- Interaction and Pathway Databases
- Laboratory Resources
- miRNA and ncRNA Databases and Tools
- Miscellaneous
- Ontology Resources
- Orthologs
- Phylogenetic Comparison Tools
- Population Biology and Polymorphism Resources
- Protein Analysis
- Public Education
- RNAi
- Sequence Analysis
- Taxonomy
- Transcription Regulation Databases and Tools
Drosophila Material Resources (Reagents)
Includes: