FlyBase:QueryBuilder Help
Query Builder Overview
QB takes advantage of how the data is stored in FlyBase to allow more sophisticated searches relative to QuickSearch or other search tools on FlyBase.
Using QB, you can search any field in a FlyBase report using a QuerySegment, and then combine the resulting hit-list with searches in other fields, allowing combinatorial searches that join QuerySegments with Boolean operators. (Note that Human Disease, Cell Line, Gene Group, and Strain reports are not currently accessible with QueryBuilder).
A set of results can be exported to QB from other searches on FlyBase, through the 'Export' button at the top of a hit-list, and then modified to refine the search by adding additional query segments.
Getting Started
Select one of the three options on the QB start page:
- Select a pre-constructed QueryTemplate
- Import a previously saved query
- Build a new query
Select a Pre-constructed Query Template
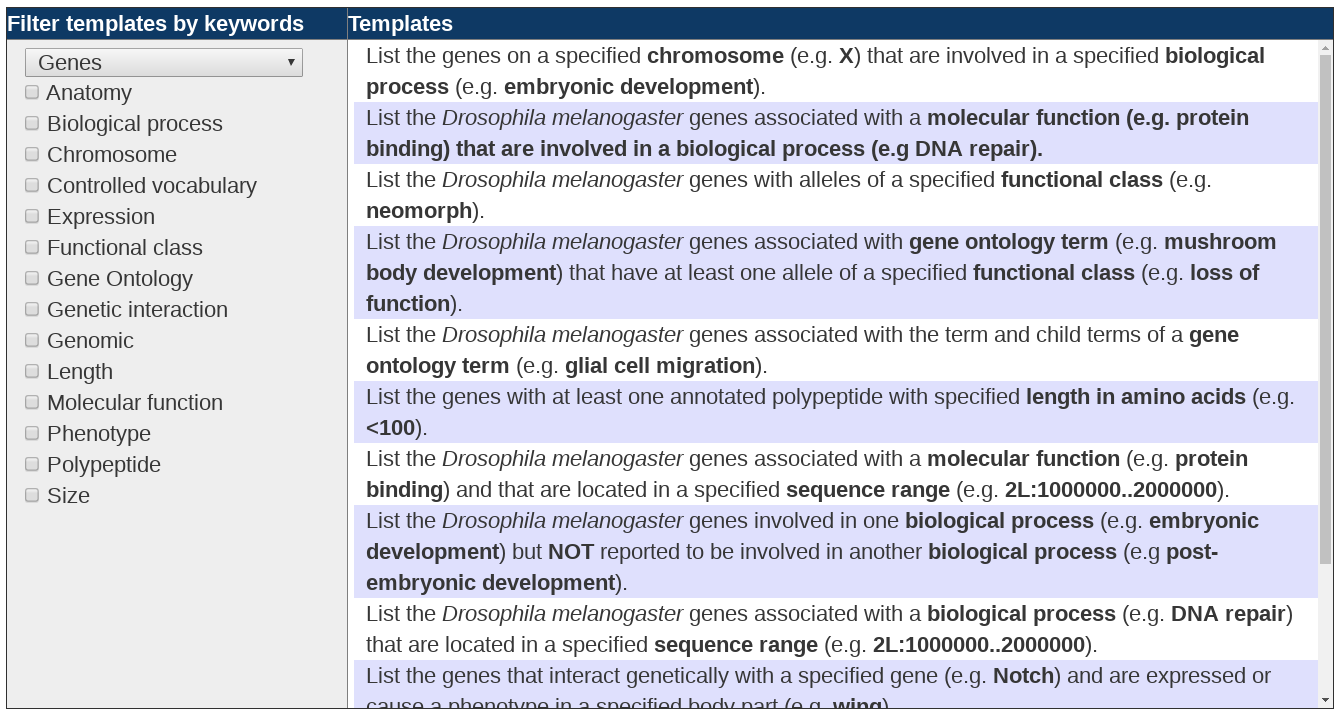
The first option on the QB start page allows one to choose a query from a large collection of pre-constructed query templates. The available templates are organized by data type. To see the list of templates related to a given class of data, choose the data class of interest from the pull down menu at the left. A list of pre-constructed query templates will appear at the right and a data class-specific list of “keywords” will appear at the left. The list of templates can be further refined by selecting one or more of the keywords. Only the templates containing the chosen keywords will remain. To return to the complete set of templates for a given data class, just deselect the chosen keywords.
Example Templates:
- List the genes associated with a specified gene ontology term (e.g. transcription factor activity) that are reported to genetically interact with a specified gene (e.g. bsk).
- List the balancers for a specific chromosome (e.g.*3LR*) available in a stock.
- List the lethal insertions for a specified gene (e.g. N).
When you find a template that matches or is similar to your query of interest, click on the template. This will bring you to a QueryBuilder Page with the specified query set up and ready to run. To modify the parameters to exactly match your own query specifications, use the green “Edit” tabs present in each segment of the query. Modify the search terms as desired, click “Finish Editing”, and then select “Run query”.
Import a Saved Query
Any QuerySchema (a collection of QuerySegments combined using Boolean operators) can be saved for running again at a later date using the “Store This Query” option on the QB results page. The QuerySchema is saved to your computer as a small text file. To run the query again, choose “Import a saved query” from the QB start page. Use the “Choose File” option to retrieve the file. The name of the chosen file will appear next to the "Choose File" box. Click on the green “Done (activate query in new QueryBuilder session)” button, which will take you to the QueryBuilder page with your saved query entered. Edit the query if desired (as described below), and then click on “Run query”.
Build a New Query
Click the yellow box on the QB start page titled' ‘Build a new query”. Follow the instructions below to either build a query using any text string of your choosing, build a query using controlled vocabulary terms, or to do an expression pattern query.
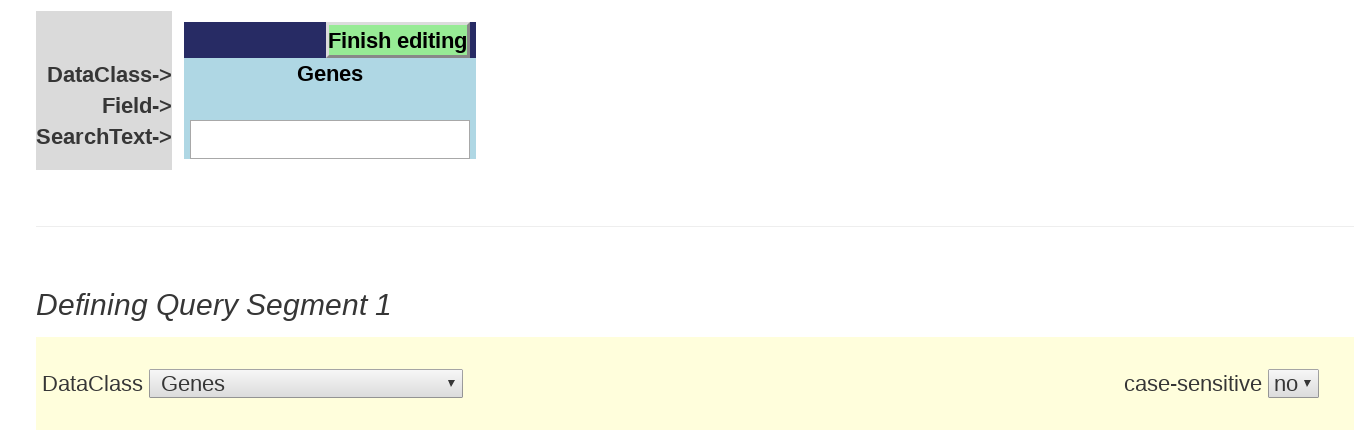
Below is a the initial Query segment box that you will see that is ready to be filled in.
Building a segment using any text string
Step 1: Select the DataClass you want to search from the DataClass dropdown menu. This will appear in the "DataClass" line of the query segment.
There are 19 options to choose from. In most cases, choosing a particular DataClass changes the window display to show the Querybuilder searchable fields found in the report for that DataSet. In other cases (Expression Search, Controlled Vocabularies) a dedicated search interface appears.
Step 2: Click on the radio button next to the report field you wish to search. This will appear in the "Field" line of the query segment.
Step 3: Enter the text string for your search in the QuerySegment’s “SearchText” box. The search algorithm will search for occurrences of the text string you entered in the specific field that you selected in step 1. In cases where the selected field value may be case sensitive (e.g. symbol), you can opt for running a case sensitive search by choosing “yes” in the “case-sensitive” dropdown menu. For some fields, autocomplete will list valid field entries guided by the text you have typed.
Step 4: Click the "Finish editing" button.
Step 5 (optional): Add an additional search segments by clicking the "+" button. A new box will appear and you can repeat the selection process. The additional segment(s) can be joined to existing segments using standard Boolean operators. The default operator is “AND”. To change to “OR” or “BUT NOT”, click on the join box until you reach the desired operator. You can remove query segments by clicking on the “x” in the top right corner of the query box.
Step 6: Click on “Run query”. (Note that the default search is for D. melanogaster. To search for results in other Drosophila species, choose the species of interest from the “Species filter” drop down menu before you run the query).
Step 7: To see results for the DataClass specified in your search, click on the appropriate green button, which indicates the number of hits and takes you to the relevant report or a hitlist. To see results in other cross-referenced DataClasses, click the green results button for the DataClass of interest.
Building a segment using a Controlled Vocabulary term
Step 1: Select "Controlled Vocabularies (CV)" from the DataClass drop-down menu.
Step 2: Clicking this option changes the window display to show top-level terms from various CVs used in FlyBase for GO ontology, anatomy, developmental stage and phenotype terms. You can either browse through the CVs from these top-level terms or you can search for terms matching what you are looking for, using the search box above the terms. By default, your search will be performed using CV terms from the whole subtree of the term you've chosen. If you wish to search only for the exact CV term you have chosen, select "This CV term only" from the “Retrieve records annotated with” drop down menu. (Hint: you'll retrieve more results by searching the whole subtree)
Step 3: Once you've decided on a term, click on the green box to use the term in your search. The window returns to the QB query page, where the first QuerySegment has been populated with your chosen CV term.
Step 4: Add additional query segments as described above (optional).
Step 5: Click on “Run query”.
Searching Gene Expression Data
Step 1: Select "Expression Patterns" from the DataClass menu.
Step 2: Build your query by entering CV terms in the Developmental Stage, body Part/Tissue, and Subcellular Location text fields. The auto-complete feature will help you choose valid CV terms to build an expression statement (see Hints and Tips).
Step 3: Click on the green 'Finish editing' button. You can edit your query before running it by clicking the green 'Edit' button, which will take you back to step 2.
Step 4: Add new clauses to your search if desired by clicking on the yellow plus sign button as described above.
Step 5: Click on the green 'Run query' button.
Step 6: Click on one of the green "Genes", "Insertions" and "Recombinant Constructs" crossreference links to get a hitlist of reports that list expression pattern data matching the search criteria for the chosen data class.
Hints and Tips for searching expression patterns:
- Note that the Expression or GAL4, etc. QuickSearch tabs provide alternative mechanisms for searching expression patterns but they do not offer the combinatorial search options of QB. In addition, the “Vocabularies” search tool (accessible from a button on the FlyBase home page) is a powerful tool for identifying CV terms of interest by providing links to vocabulary term reports that include term definitions, hierarchical CV term tree structures, and other useful data.
- The input fields in the QB expression pattern form use a sophisticated auto-complete feature. When you begin typing in (or even just click inside) a field, a list of suggested CV terms will appear. For the first field you fill in, all appropriate CV terms for that category are available.
- Each filled search field further constrains the auto-complete function for the remaining fields. For example, if you have entered "gastrula stage" in the Developmental Stage field, the auto-complete function for the Body Part/Tissue search field will include the CV term "parasegment 10", but will exclude the CV term "leg". Likewise, if you have entered the CV term "prothoracic leg" in the Body Part/Tissue seach field, the auto-complete function for the Developmental Stage search field will include "adult stage" but exclude "embryonic stage 4".
- If you select only terms suggested by the auto-complete feature, your expression statement query should always match some results. To avoid running queries which produce no hits, it is highly recommended that you use terms suggested by the auto-complete feature.
- Below each search field is a Qualifier field, in which you can enter a qualifier, such as "early" for Developmental Stage, or "apical" for Subcellular Location. Each of the qualifier search fields also has an auto-complete function, and will only offer qualifiers that have been used in curation with the term entered in the search field above it.
- The auto-complete cannot take into account that an expression statement may only exist in, e.g., the "Insertions" dataset, when you are currently searching the "Genes" dataset. In these cases, your search will return no direct hits, but the green "Cross references" buttons that appear when you run the query indicate that there are hits in another dataset
Additional QB Features
Wild Cards
The Asterisk is wild. An asterisk (*) on either end of your search string, or embedded in the middle of the string, is interpreted as "any character". Wild cards are not automatically added to QB searches. If a query is unproductive, try it again with * on one or both ends.
- Stocks | FlyBase Genotype mam*
- Alleles | Phenotypic Class *maternal*
- Insertions | Symbol *ptc*
Any value, no value
Search for the presence or absence of information in a field, rather than a specific value.
The options are IS NULL and IS NOT NULL (this query is case sensitive).
Logical operators
Combine multiple query legs with logical operators.
The options are AND, OR, and BUT NOT.
When using two or more query segments, QB gives precedence to the previous segments.
- haltere AND wing OR leg is interpreted as (haltere AND wing) OR (leg)
Phrases
Multiple words are treated as a phrase.
Only records that include the search words in the order you specify will be matched.
Calculations
Calculations can be incorporated into searches of some fields that contain numbers.
The options are greater than (>), less than (<), plus or minus (+/-) and range (-).
- Polypeptides | Length (aa) | <50
- Polypeptides | Predicted MW | <25
- Transcripts | Length (nt) | 100-200
- Genes | Number of transcripts | >10
Hierarchical CV queries
The GO and Anatomy/Development term relationships are fully supported in QueryBuilder.
Searches of CV fields within standard data classes (e.g., Genes) find only records that contain the individual term you specify. The GO/Anatomy CV database associates each term in these CVs with all of the terms below it in the hierarchy, allowing a single search to find records that contain a term or any child of that term.
Case Sensitivity
Case-insensitive searches are standard. There are two exceptions:
- A case-sensitive Symbol search is available for most data classes.
- The reserved phrases IS NULL and IS NOT NULL are case sensitive.
Cytological Location Searches
Cytological Location Searches are redirected to the GBrowse dataset, which uses estimated sequence ranges of cytological locations.
Controlled Vocabularies
To access all of the Gene Ontology, Anatomy, Developmental stage, and Phenotype Controlled Vocabularies (CVs) used in FlyBase, go to Vocabularies.
Field content dictionaries
Preview the information in a field, or select dictionary entries to use in a search.
The field dictionary lists up to 100 most-commonly-used symbols, terms, numbers or words from the data in the selected field. Access the dictionary by clicking on the yellow box in the upper right labelled “select search text from dictionary”. Choose the term of interest and click on “use selected content”.